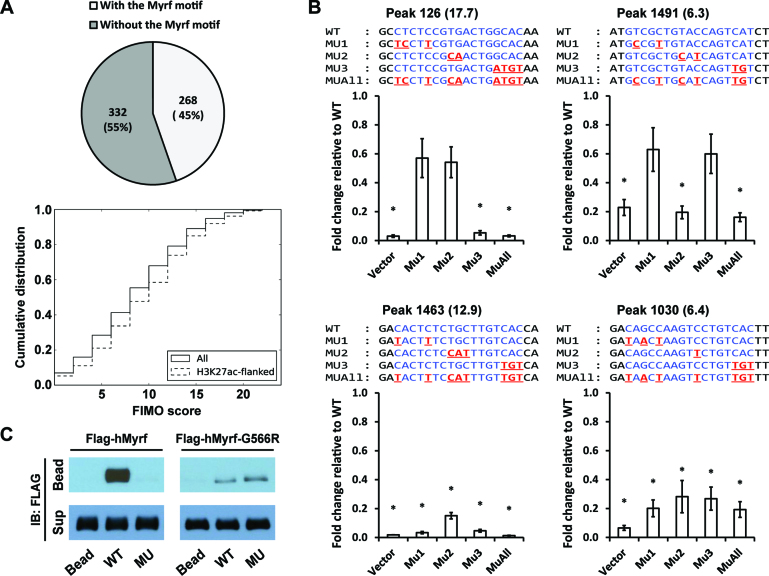
Figure 6.
Detailed analysis of the Myrf motif. (A) MEME randomly chose 600 Myrf ChIP-seq peaks from a total of 2052 for its run and found 268 of them to have an incidence of the Myrf motif. As a further way of evaluating the prevalence of the Myrf motif, FIMO was used to scan the 2052 Myrf ChIP-seq peaks, searching for matches of the Myrf motif. Motif matches were ranked by the FIMO score, and a cumulative distribution was computed. (B) Luciferase assay was performed for an in-depth analysis of the Myrf motif. Four motif matches were cloned into pGL3-promoter. The rn4 genomic coordinates are as follows: 126 (chr1:169920248–169920647), 1463 (chr4:123025896–123026295), 1030 (chr2:28381432–28381831) and 1491 (chr4:147568152–147568551). The numbers in parentheses are FIMO scores. For each motif match, the first, second, and third unit was selectively mutated in MU1, MU2 and MU3, respectively. MUAll combines the mutations of MU1, MU2 and MU3. For all samples, mMyrf-expressing plasmids and pRL-TK were co-transfected. pGL3-promoter was used as a control (Vector). The transcriptional readout from the WT sequence was set to 1, and the reported values are means and standard errors of 4 biological replicates. *P < 0.05 by two-tailed one sample Student's t test with Bonferroni correction. (C) DNA pulldown assay using duplex oligonucleotides corresponding to the Myrf motif incidence shown in Figure 5B. The N-terminal fragments of Flag-hMyrf-G566R (which exist as monomers) bound to WT, but very weakly. Further, they failed to distinguish between WT and MU, unlike their homo-trimeric counterparts. IB: immunoblotting.