Figure 3.
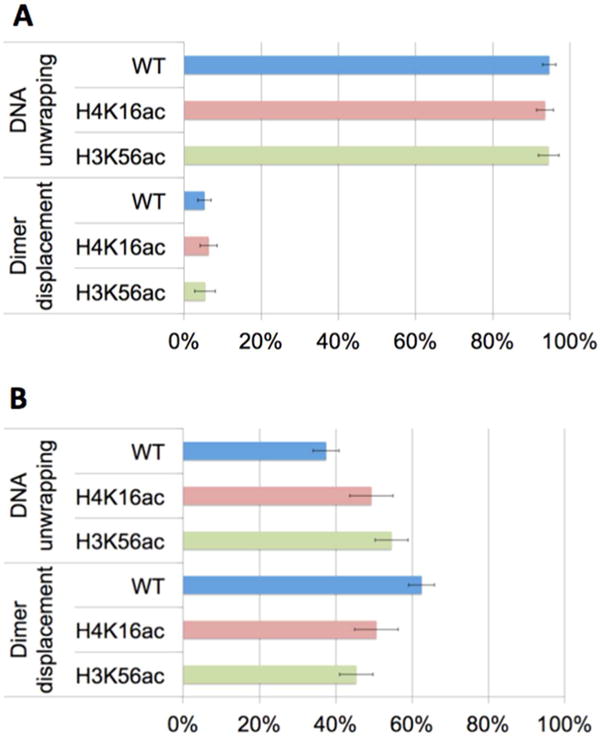
Population densities of nucleosomes grouped by the initial dimer dissociation step. The two populations are denoted by “DNA unwrapping” and “Dimer displacement”. Nucleosomes in the “DNA unwrapping” group start dimer dissociation by DNA unwrapping prior to dimer displacement. Nucleosomes in the “Dimer displacement” group start dimer dissociation by dimer displacement prior to DNA unwrapping. Dimer displacement was induced by (A) a high salt level and (B) Nap1. WT, H4K16ac, and H3K56ac denote wild-type nucleosome (i.e., no acetylation), nucleosome acetylated at H4K16, and nucleosome acetylated at H3K56, respectively. The error bars represent the standard deviations of the counts assuming a binomial distribution whose variance is given by np(1 − p), where n is the sample size and p is the probability. (A) In the 95% of the population, dimer dissociation induced by salt starts with DNA unwrapping and the effect of acetylation on this statistics is negligible. (B) In the 63% of the population, dimer dissociation mediated by Nap1 starts with dimer displacement (wild-type, WT). This population density decreases significantly upon histone acetylation at H4K16 or H3K56 (Table 1C).
