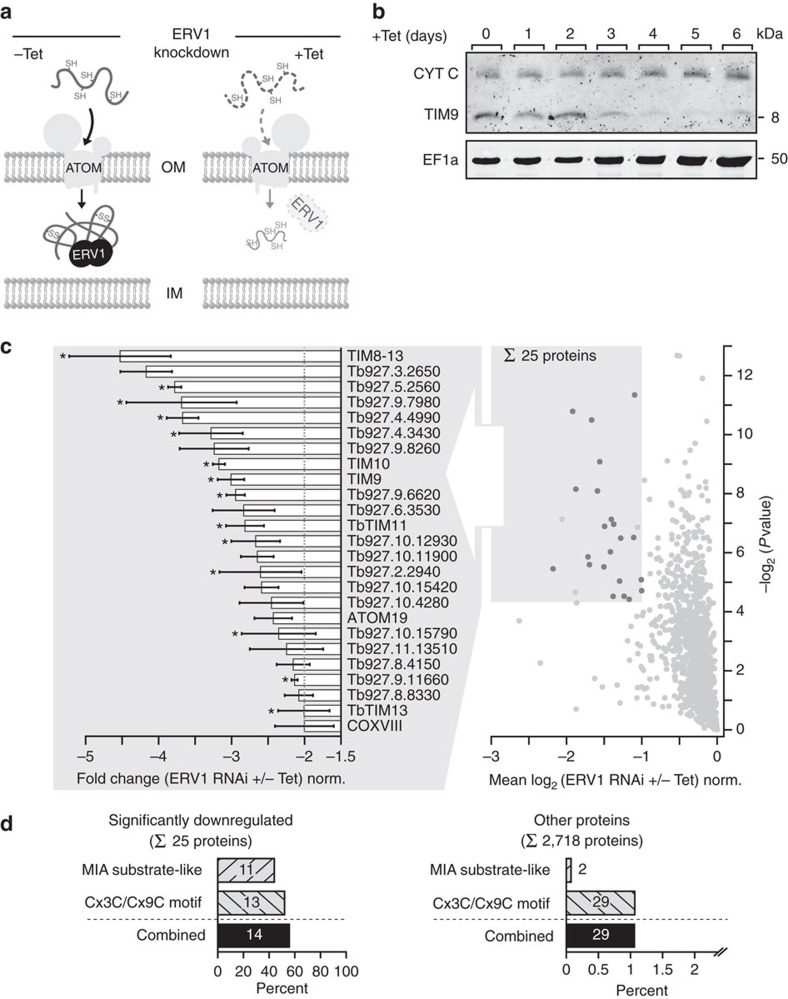
Figure 5. Identification of putative MIA pathway substrates by ablation of the sulfhydryl oxidase ERV1.
(a) A subset of nuclear-encoded soluble proteins located in the mitochondrial intermembrane space (IMS) is imported via the mitochondrial IMS assembly (MIA) pathway. Import and correct folding of these cysteine-rich proteins requires the sulfhydryl oxidase ERV1 (−Tet). Tetracycline-induced RNAi-mediated knockdown of ERV1 results in impaired import and depletion of MIA pathway substrates in the IMS (+Tet) and their subsequent degradation in the cytosol. (b) Immunoblot analysis of whole-cell extracts showing the steady-state levels of the IMS proteins cytochrome c (CYT C) and TIM9 as well as the cytosolic protein EF1a following tetracycline-induction (+Tet) of ERV1 RNAi for the indicated time. (c) Proteins reduced in abundance in mitochondria-enriched fractions of induced ERV1-RNAi cells. MS-based quantification of proteins from induced (+Tet) versus uninduced (–Tet) cells was based on peptide stable isotope dimethyl labelling (n=3). Proteins with a mean log2 ratio of less than or equal to −1 (corresponding to a fold change of less than or equal to −2) and a P value <0.05 (shaded area) were considered significantly reduced in abundance on ERV1 knockdown. Right graph: dark grey dots, significantly reduced proteins also present in the mitochondrial importome. Left graph: error bars indicate the s.e.m.; *predicted MIA pathway substrates. (d) Putative MIA pathway substrates predicted by similarity to known MIA substrates in other organisms (MIA substrate-like) and a general Cx3C/Cx9C motif-based approach are highly overrepresented among the set of proteins significantly downregulated following ERV1 knockdown (left) compared to the set of all other proteins quantified in this experiment (right). Shown is the percentage of putative MIA pathway substrates predicted by the aforementioned approaches in each subset.