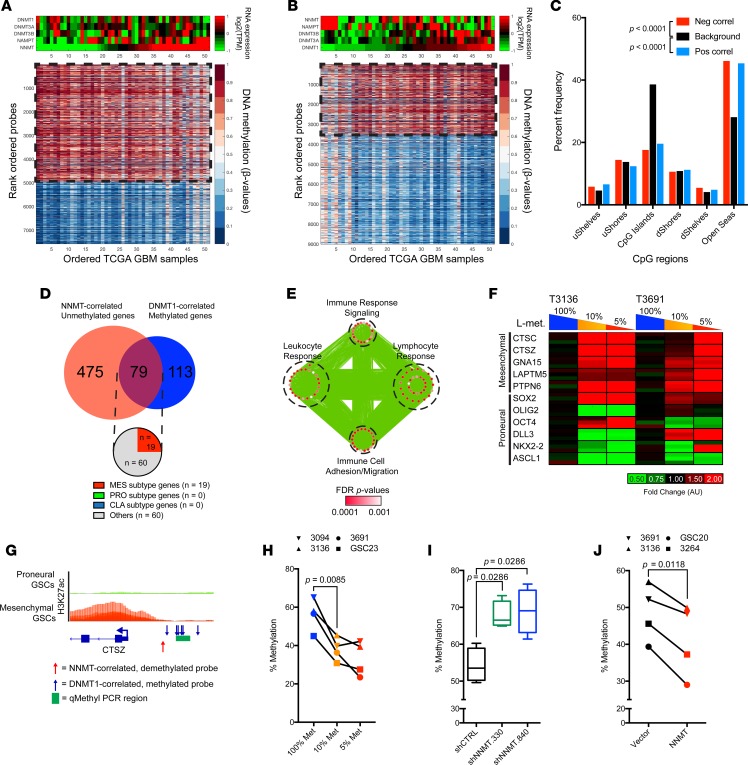
Figure 7. NNMT and DNMT1 affect DNA methylation of inflammatory genes.
(A) CpG probes significantly anticorrelated with NNMT mRNA expression in TCGA DNA methylation 450K array and RNAseq datasets (n = 51). (B) CpG probes with significant positive correlation with DNMT1 mRNA expression in TCGA DNA methylation 450K array and RNAseq datasets (n = 51). (C) Distribution of CpG region in probes with significant positive or negative correlation with NNMT expression (pos correl; neg correl) compared with nonsignificantly correlated CpG probes. Differences in distribution tested with 2-way ANOVA. (D) Annotated genes with significant negative correlation with NNMT mRNA expression and significant positive correlation with DNMT1 mRNA expression. (E) Gene ontology analysis of the common genes among those with significant negative correlation with NNMT mRNA expression and significant positive correlation with DNMT1 mRNA expression. Size of each node indicates the size of each gene set; thickness of each edge corresponds with the number of genes shared between connected nodes. (F) RT-PCR analysis of mesenchymal subtype genes in proneural T3136 and T3691 cultured in different methionine restriction conditions (100% Met, 115.5 μM; 10% Met, 11.5 μM; 5% Met, 5.75 μM). (G) Schema for methylation-sensitive (qMethyl) RT-PCR. Blue arrows indicate CpG probes significantly correlated with DNMT1 expression. Red arrow indicates CpG probe significantly correlated with NNMT expression. H3K27ac ChIP-seq data of proneural and mesenchymal GSCs (Stephen C. Mack, unpublished observations) visualized on IGV 2.3.80. (H) Quantification of DNA methylation levels of CpG loci in the CTSZ promoter region in 4 proneural GSC models (T3094, T3136, T3691, GSC23) cultured in different methionine restriction conditions (100% Met, 115.5 μM; 10% Met, 11.5 μM; 5% Met, 5.75 μM). Two-tailed Student’s t test used to determine changes in mean between conditions. (I and J) Quantification of DNA methylation levels of CpG loci in the CTSZ promoter region in 2 proneural GSC models (T3136 and T3691) and 2 mesenchymal GSC models (T3264 and GSC20) (I) transduced with nontargeting control or shRNA clones targeting NNMT or (J) transduced with vector control or NNMT overexpression constructs.