Figure 5. The novel DHBV RNA species is protected from MNase digestion.
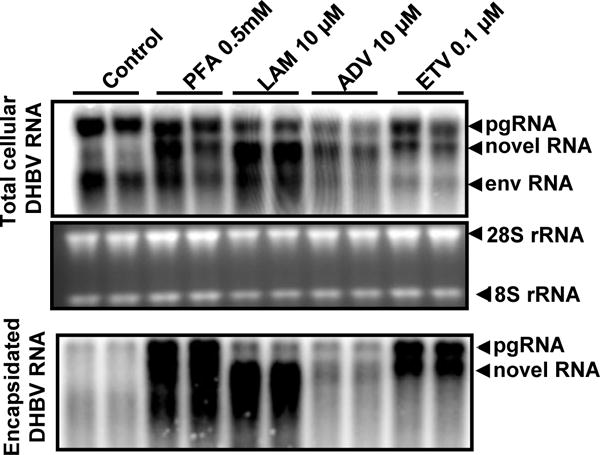
dstet5 cells were left untreated (Control) or treated with the indicated DNA polymerase inhibitors for 3 days in the absence of tet. Cell lysates were centrifugated at 10,000 g for 10 min to remove cell debris and nuclei. The supernatants were digested with 20 U/ml of MNase at 37°C for 30 min. The core particles were precipitated with 35% PEG 8000 in ice for 60 min. Encapsidated RNA was extracted with TRIzol reagent. Total cellular RNA (upper panel) and encapsidated DHBV RNA (lower panel) were analyzed by Northern Blot hybridization. Ribosomal RNAs (28S and 18S rRNA) served as loading controls. The two lanes under each treatment condition are the results of experimental duplicates. The positions of DHBV pgRNA and subgenomic mRNA encoding viral envelope proteins (envRNA) are indicated. The RNA species accumulated in the cells treated with DNA polymerase inhibitors are indicated as novel RNA.
