Figure 7. Accumulation of the novel RNA species requires RNase H activity.
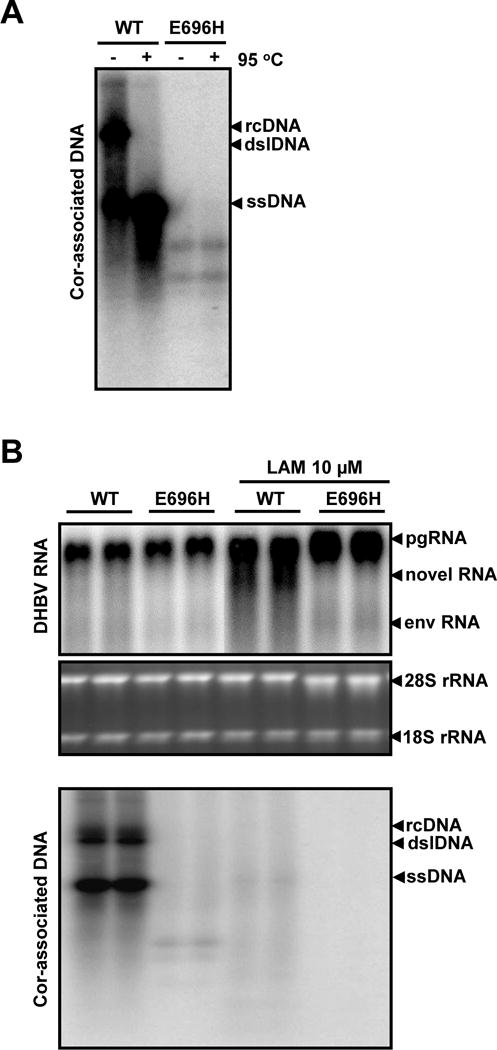
Chicken hepatoma cells (LMH) were transfected with plasmid pCMV-DHBV (WT) or pCMV-DHBV/PE696H (E696H). (A) Forty-eight hours post transfection, the core-associated viral DNA were extracted, resolved in agarose gel, without or with denaturation in 50% formamide at 95°C before loading as indicated on the top of the figure, and detected by hybridization with a full-length riboprobe-specific for minus-stranded viral DNA. (B) The transfected cells were left untreated or treated with 10 μM of lamivudine for 48h. Total cellular RNA and core-associated DNA were extracted and analyzed by Northern (upper panel) and Southern (lower panel) hybridization, respectively. For RNA analysis, ribosomal RNAs (28S and 18S rRNA) served as loading controls. The positions of DHBV pgRNA and subgenomic mRNA encoding viral envelope proteins (envRNA) are indicated. The RNA species accumulated in the cells treated with LAM are indicated as novel RNA. For core-associated DHBV DNA, the forms of relaxed circular (RC), double stranded linear (DSL) and single stranded (SS) DNA are indicated.
