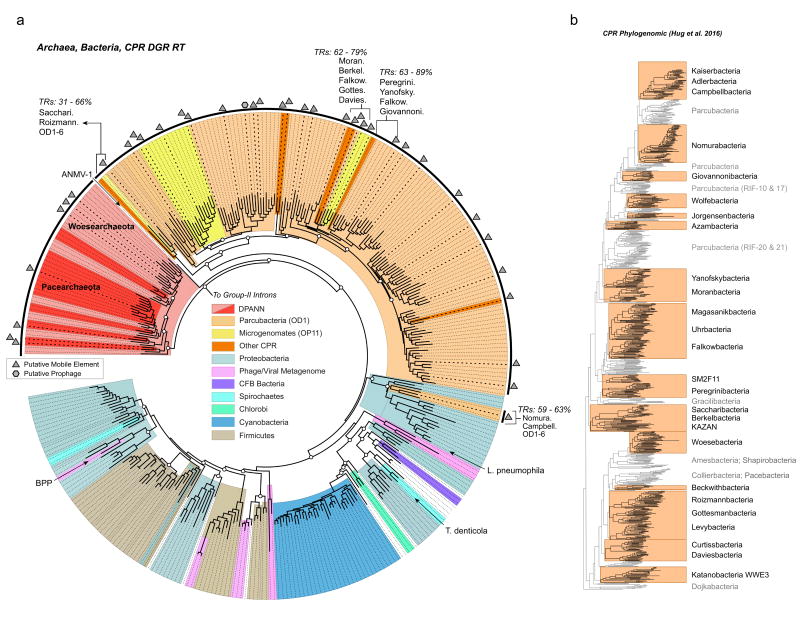
Figure 2. Phylogeny of diversity-generating retroelements and radiation of novel lineages.
a, Phylogeny and taxonomic association of diversifiers. Sequences obtained in this study are highlighted in black on the outer edge of the tree. Shaded slices indicate either candidate superphyla comprising groundwater organisms, or bacterial phyla and phage with previously sequenced RTs. The tree was constructed with 346 sequences and 261 alignment positions. Archaeal sequences from the DPANN superphylum are indicated in either dark red (Pacearchaeota), or light red (Woesearchaeota) shaded slices. Paraphyletic groups of species with closely related RTs are indicated with the associated range of pairwise TR sequence similarity. Symbols (hexagons and triangles) highlight DGRs identified in this study, which are found in close proximity to putative prophage or mobile elements (i.e. transposons or conjugative elements). Diversifiers that have been previously examined are: BPP, a Bordetella pertussis phage; T. denticola, Treponema denticola; L. pneumophila, Legionella pneumophila strain Corby; and ANMV-1, a marine virus of an uncultivated archaeal host. White circles indicate bootstrap values >70% for basal nodes. The scale indicates amino acid substitutions per site. b, Phylogenomic tree of CPR organisms, highlighting major lineages that contain at least one DGR. The phylogenomic tree was originally presented by Hug et al. (2016).