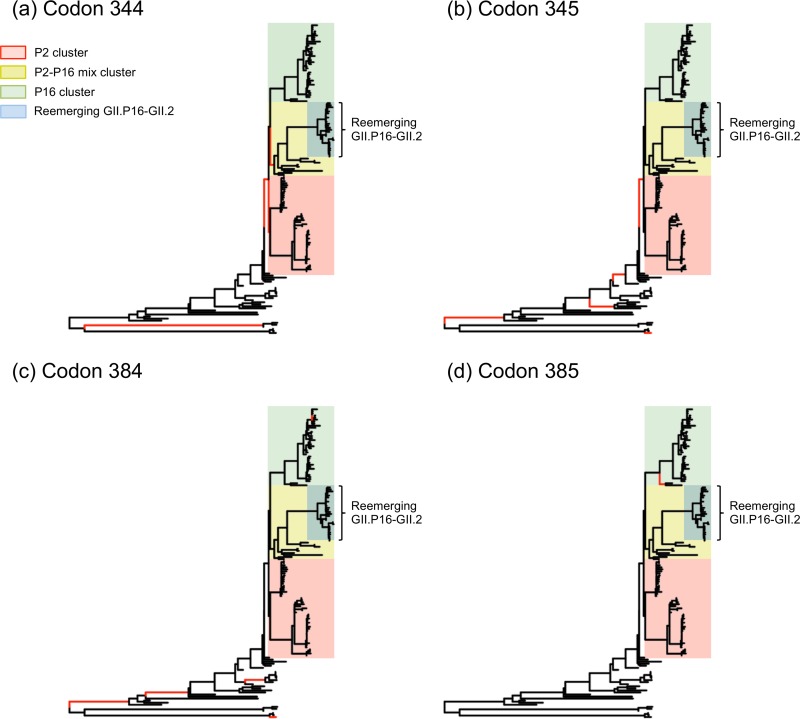
FIG 3 .
Positive evolution of the major capsid protein of GII.2 noroviruses. Maximum likelihood trees of VP1 nucleotide (nt) sequences of GII.2 indicate the branches under possible positive selection. The branches with evidence of positive selection (empirical Bayes factor of >10, P < 0.05; estimated using the mixed-effect model of evolution [MEME] method) at codon positions 344 (a), 345 (b), 384 (c), and 385 (d) are represented by red branch lines. The color shade indicates the phylogenetic clustering based on the polymerase genotype as indicated in Fig. 1. Reemerging GII.P16-GII.2 strains are indicated in blue.