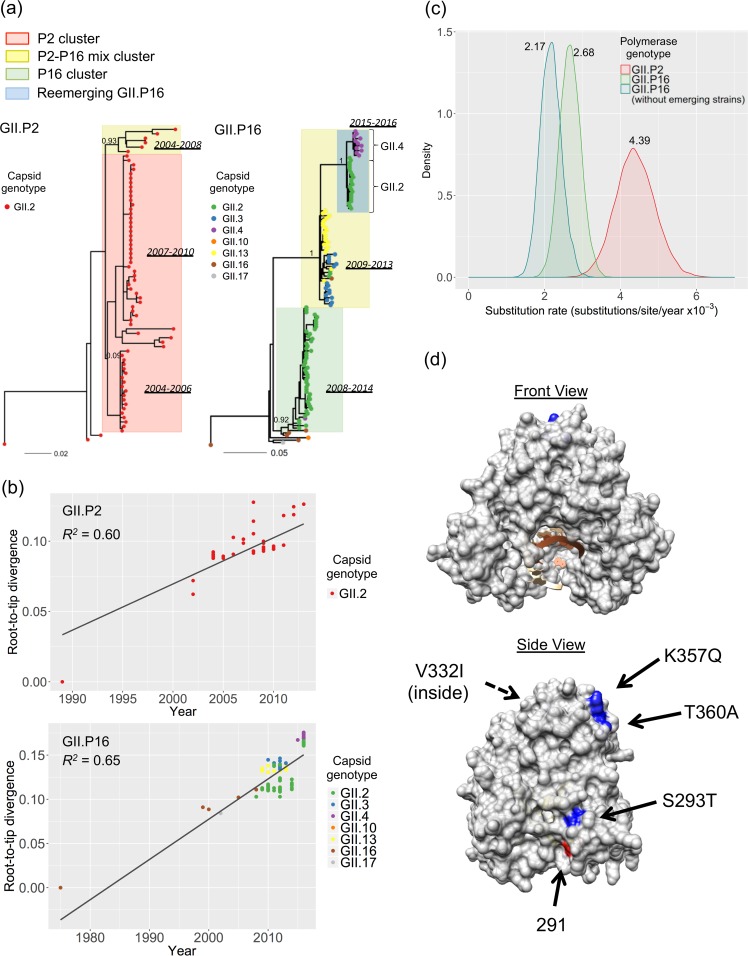
FIG 4 .
Evolutionary dynamics of the GII.2 polymerase. (a) Maximum likelihood trees of RdRp-encoding (nt) sequences of strains GII.P2 (n = 72) and GII.P16 (n = 131). Phylogenetic clusters based on VP1 nt sequences (Fig. 1) are shown here; the P2 cluster is represented in red, the P16 cluster in green, and the P2-P16 mix cluster in yellow. Reemerging GII.P16 strains, including GII.P16-GII.2 and GII.P16-GII.4, are indicated in blue. Each strain is represented by a circle, with different colors according to the associated capsid genotypes; GII.P2-GII.2 and GII.P16-GII.2 strains are indicated by red and green, respectively. The numbers on the ancestral nodes of the clusters represent the node support value calculated by the approximate likelihood-ratio test using PhyML. (b) The root-to-tip divergence plot of RdRp-encoding (nt) sequences of GII.P2 and GII.P16 strains. The x axis indicates isolation year, and the y axis shows the root-to-tip divergence on the maximum likelihood phylogenetic tree. The black lines indicate a linear regression line of the root-to-tip divergence and isolation year. Each strain is colored according to its capsid genotype. The genomic regions used spanned nt 4385 to 5104 relative to Snow Mountain virus (GenBank accession number AY134748). (c) The kernel density plot indicates the posterior estimates of substitution rate (substitutions/site/year) among GII.P2, GII.P16, and GII.P16 without reemerging strains. (d) Structural mapping of the amino acid (aa) substitutions in the reemerging GII.P16-GII.2 strains. Conservative mutations in the RdRp aa sequence from the reemerging GII.P16 strains compared to pre-2016 GII.P16 strains (P16 cluster and P2-P16 mix cluster) are mapped on the structural model of a GII.P4 RdRp (PDB number 4QPX). The molecular model was visualized using Chimera v.1.11. aa substitutions are indicated in blue. Residue 291, a residue that was shown by Bull et al. (46) to alter the incorporation rate of GII.4 polymerases, is indicated in red.