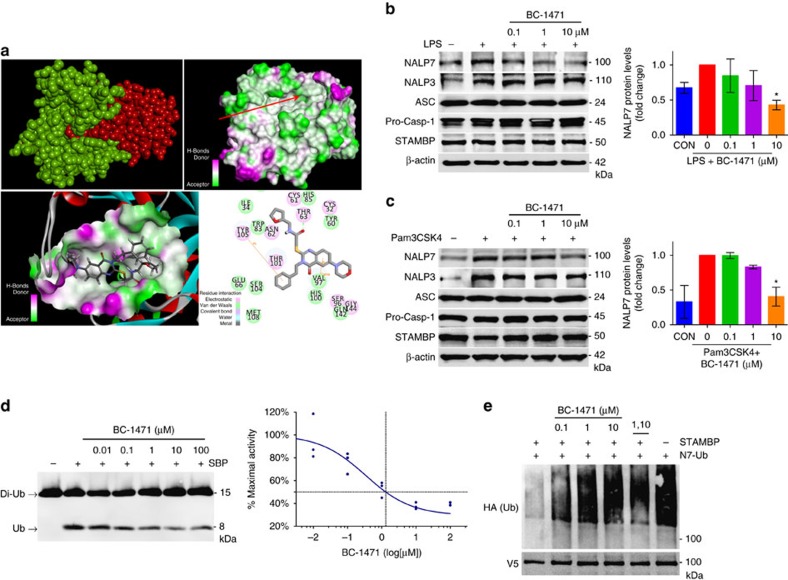
Figure 5. STAMBP inhibition with small molecule BC-1471 destabilizes NALP7 protein.
(a) Top, left, a structural model of STAMBPL1 (green) interacting with Ub (red). Top, right, molecular charge mapping with red arrow pointing to the Ub-binding groove of STAMBP. Bottom, left, topological docking model of BC-1471 in the Ub-binding groove of STAMBP. Bottom, right, 2D mapping of key putative contact sites driving STAMBP and BC-1471 interaction. Amino acid numbers correspond to the residues of the published crystal structure of the catalytic domain of STAMBP (3RZU.pdb). (b,c) BC-1471 pre-treatment decreased NALP7 protein abundance after 6 h exposure to (b) LPS (200 ng ml−1) or (c) Pam3CSK4 (100 ng ml−1) compared to vehicle control (DMSO) in THP-1 cells. Right, densitometric analysis of the NALP7 signal for each condition relative to the treated positive control. Data shown as mean±s.d. (n=3). *P<0.05. (d) In vitro DUB assay with K63-linked di-Ub. BC-1471 inhibited cleavage of K63-linked di-Ub (200 nM) to mono-Ub by purified recombinant STAMBP (25 nM) incubated at 37 °C for 2 h in a concentration dependent manner. Graph depicts an inhibitor–response curve with IC50 determined by non-linear regression (n=3). (e) In vitro DUB with NALP7 substrate. Immunopurified Ub-NALP7 substrate abundance was preserved with BC-1471 (10 μM) and 1,10-phenanthroline (5 mM) treatments, compared to vehicle control when incubated at 37 °C for 60 m with purified recombinant STAMBP (1 μM). V5 is shown as a loading control. (b,c) Two-way analysis of variance (ANOVA) with post hoc Dunnett's multiple comparisons test.