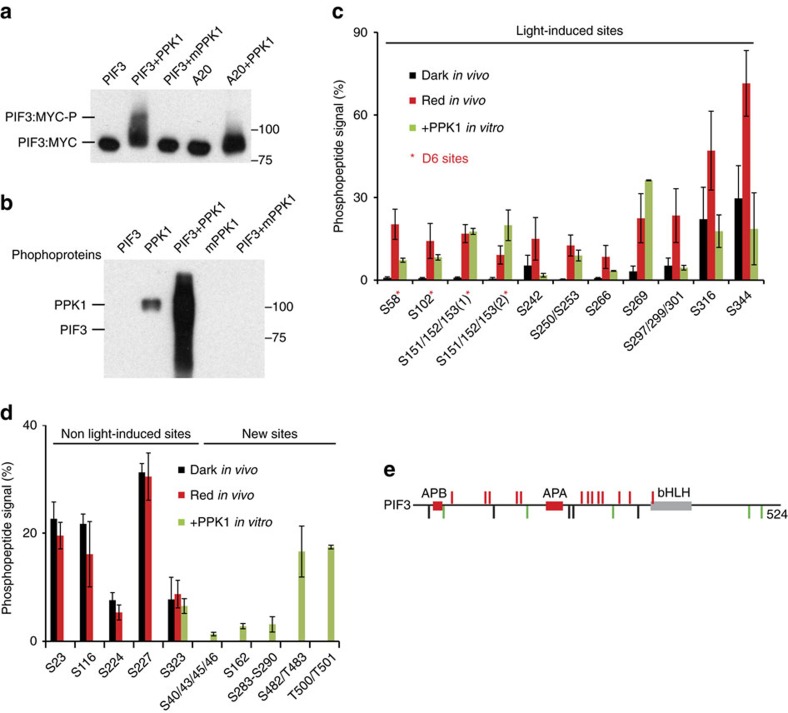
Figure 4. PPK1 phosphorylates PIF3 at light-inducible phosphosites in vitro.
(a) PPK1 induces a strong mobility shift in PIF3 in an in vitro kinase assay. PPK1 and MYC-tagged PIF3 variants, affinity-purified after expression in E. coli, were combined under protein-kinase-assay conditions and examined for an induced, potentially phosphorylation-related, mobility-shift in PIF3 by immunoblot blot analysis using anti-MYC antibody. mPPK1: kinase-dead mutant of PPK1; A20: phospho-dead mutant of PIF3 mutated in the 20 phosphoresidues induced by light in vivo. (b) PPK1 phosphorylates PIF3 in vitro. Phosphoproteins from the indicated in vitro protein-kinase-assay combinations were detected by western blot using pIMAGO-biotin. (c,d) Mass-spectrometric analysis of in vitro, PPK1-catalysed phosphosites in PIF3 compared to those sites established as rapidly induced (c), or unaffected (d), by red light (Rp) in vivo. d (left), constitutively phosphorylated, non-light-induced in vivo, (right) not detectably phosphorylated in dark or light in vivo, PPK1-induced in vitro. Phosphopeptide Signal (%) corresponds to the percent of the residues at each site that are phosphorylated in Dk-grown or Rp-treated seedlings, or in PPK1-treated PIF3 in vitro. D6, strongly light-induced sites in vivo. Data are the means of biological repeats±s.e. (e) Schematic depiction of PIF3 phosphosites. Red bars represent phosphosites that are both light-induced in vivo and catalysed by PPK1 in vitro. Black bars represent sites that are constitutively phosphorylated in vivo, but not detectably phosphorylated by PPK1 in vitro, except S323. Green bars represent newly identified phosphosites catalysed by PPK1 in vitro. APB, active phyB-binding domain; APA, active phyA-binding domain; bHLH, basic helix-loop-helix domain.