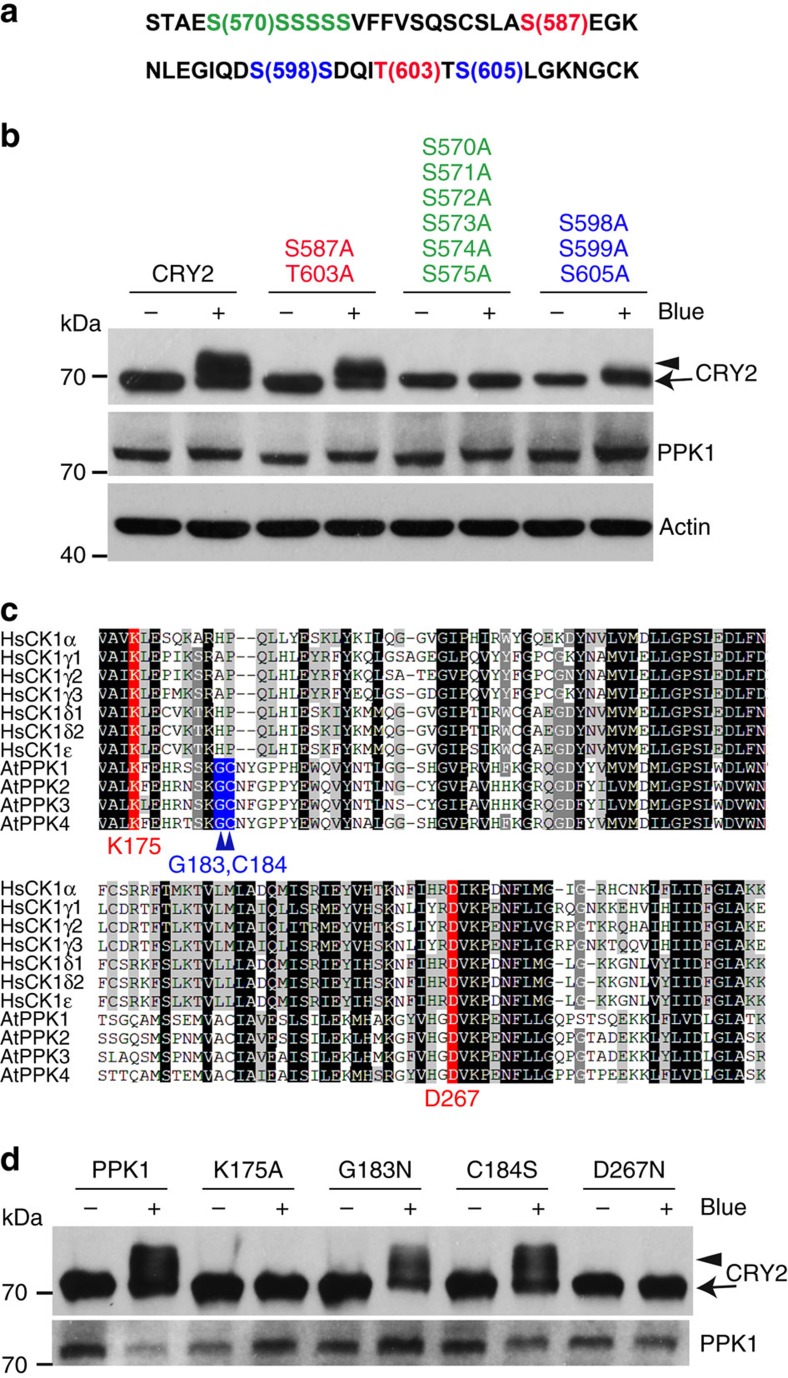
Figure 7. Mutation analyses of PPK1-catalysed and blue light-dependent phosphorylation of CRY2.
(a) A partial amino acid sequence in CRY2, in which the phosphosites subject to site-specific mutations are highlighted by colours. (b) Immunoblot showing PPK1-catalysed phosphorylation of CRY2 and three indicated CRY2 mutants corresponding to the phosphosites shown in a, including CRY2S587A/T603A, which was the proposed CK1.3 and CK1.4 phosphorylation sites, CRY2S570A/S571A/S572A/S573A/S574A/S575A, which contains mutations in the serine cluster (S570–S575), and CRY2S598A/S599A/S605A, which contains mutations in three serine residues (S598, S599 and S605) that are the major phosphosites of CRY2 detected (Fig. 1b). Cells transfected to co-express CMV::Myc-CRY2 or CRY2 mutants and CMV::Flag-PPK1 were kept in the dark (Blue −) or exposed to the blue light (Blue +; 30 μmol m−2 s−1) for 60 min. CRY2 and PPK1 were detected by immunoblot probed with anti-Myc or anti-Flag antibodies, respectively. (c) Amino acid sequence alignment of the kinase domain in Arabidopsis PPKs and human CK1s. The two evolutionarily conserved residues (K and D, including K175 and D267 of PPK1) are labelled in red; the other two residues that are conserved only in Arabidopsis PPKs (G and C, including G183 and C184 of PPK1) but not in CK1 are labelled in blue. (d) HEK293T cells co-expressing CMV::Myc-CRY2 and CMV::Flag-PPK1 or the CMV::Flag-PPK1 mutants were treated with blue light and analysed by immunoblot probed by anti-CRY2 or anti-Flag, respectively.