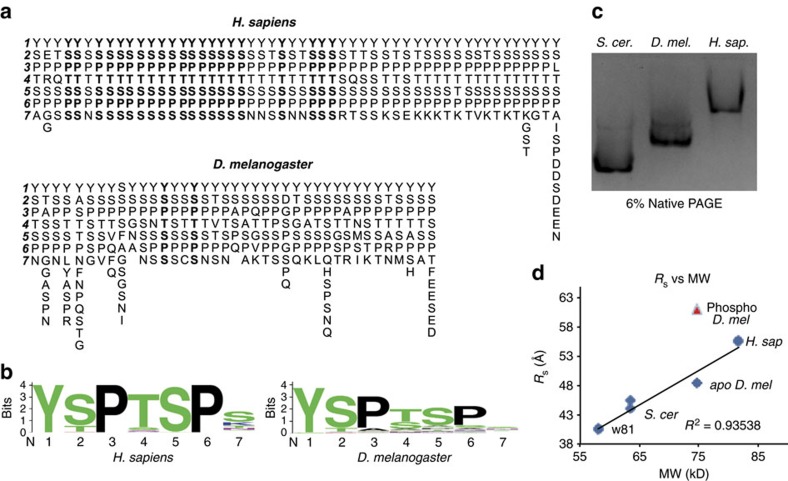
Figure 1. A similar global CTD structure exists despite sequence differences.
(a) The sequences of the CTDs of D. melanogaster and H. sapiens with each repeat beginning with YSP arranged vertically and the CTD oriented from left to right. Consensus heptads are in boldface. The Drosophila CTD has only two consensus heptads. (b) Motif conservation of fly and human CTDs. (c) Native gel electrophoresis of the S. cerevisae, D. melanogaster and H. sapiens MBP-CTD fusion proteins, with pIs of 5.57, 5.83, 5.93, respectively. Electrophoretic mobility scales with CTD length, suggesting structural homology. (d) RS versus molecular weight (MW) derived from size exclusion chromatography analysis of MBP-CTD fusions. Two replicates of each protein are plotted (several points appear as one because of overlapping values). RS is linearly related to MW, suggesting gross structural homology. Phosphorylation of the D. melanogaster CTD by P-TEFb increases RS, causing the CTD to deviate from the line (red triangles).