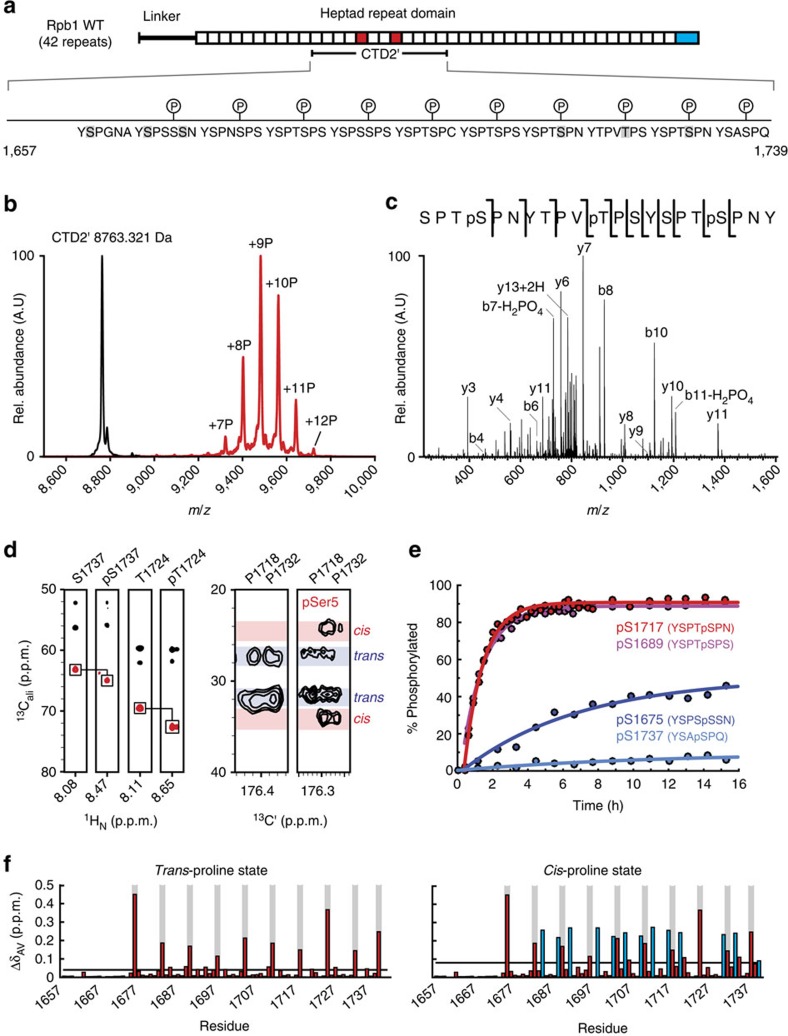
Figure 2. CTD2′ Ser5 phosphorylation probed by MS and NMR spectroscopy.
(a) Amino-acid sequence of CTD2′ displaying 99% confidence phospho-site assignments by MS/MS (grey boxes) and by NMR (P-symbols). MS/MS peptide coverage was complete. (b) Linear positive MALDI TOF MS of unphosphorylated CTD2′ (black) and hyper-pSer5 CTD2′ (red). (c) Representative spectrum from Nano-LC MS/MS analysis of hyper-pSer5 CTD2′. (d) Representative strips from 3D HNCACB spectra of unphosphorylated and hyper-pSer5 CTD2′ showing perturbation upon phosphorylation (left) and strips from 3D CCCON spectra of unphosphorylated and hyper-pSer5 CTD2′ showing pSer5-induced trans-to-cis isomerization of Pro6 (P1718 and P1732; right). (e) Representative kinetic traces for CTD2′ phosphorylation monitored by RT-NMR. (f) Average chemical shift perturbations for CTD2′ upon phosphorylation for the trans-proline-enriched (red) and cis-proline-enriched states (blue). Grey bars indicate pSer/pThr5 residues and the black line denotes the average perturbation.