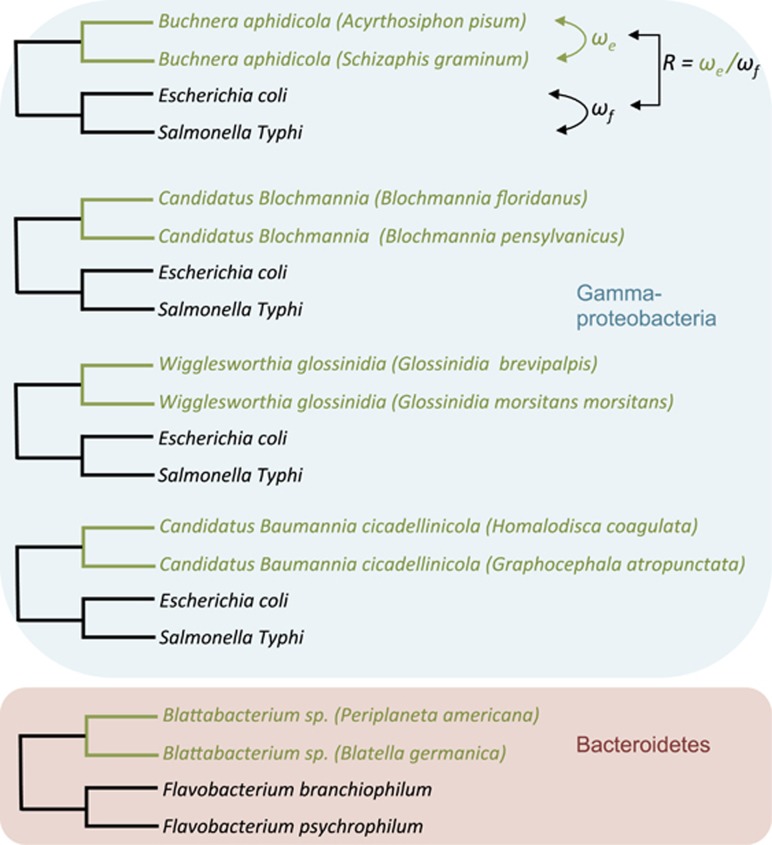
Figure 1.
Determining the relative strength of selective constraints in endosymbiotic bacterial genomes. We calculated the ratio between non-synonymous nucleotide substitutions per non-synonymous site (dN) and nucleotide substitutions per synonymous site (dS) (ω=dN/dS) to estimate the strength of selection on protein-coding genes. This ratio was estimated between pairs of endosymbiotic genomes within each of the five endosymbiotic systems (ωe) and between pairs of relative free-living bacteria (ωf). We used as endosymbiotic gamma-proteobacteria: (1) Buchnera aphidicola (strains from aphids Acyrthosiphon pisum and from Schizaphis graminum), (2) Candidatus Blochmannia (Blochmannia floridanus and Blochmannia pennsylvanicus), (3) Wigglesworthia sp. (Wigglesworthia glossinidia and Wigglesworthia morsitans) and (4) Candidatus Baumannia cicadellinicola (Homalodisca coagulata and Graphocephala atropunctata). We used as endosymbiotic bacteria from the Bacteroidetes group Blattabacterium sp. (Blattabacterium from Blatella germanica and from Periplaneta americana). We used Escherichia coli and Salmonella enterica as the external free-living bacteria pair relatives of gamma-proteobacteria and Flavobacterium branchiophilum and F. psychrophilum as free-living bacterial relatives of Bacteroidetes endosymbionts. We analyzed how the selective constraints on genes varied when comparing endosymbiotic bacteria (in green) with their free-living relatives (in black). The selective constraints on endosymbiotic bacterial genomes relative to their free-living bacterial relatives was calculated as R=ωe/ωf.