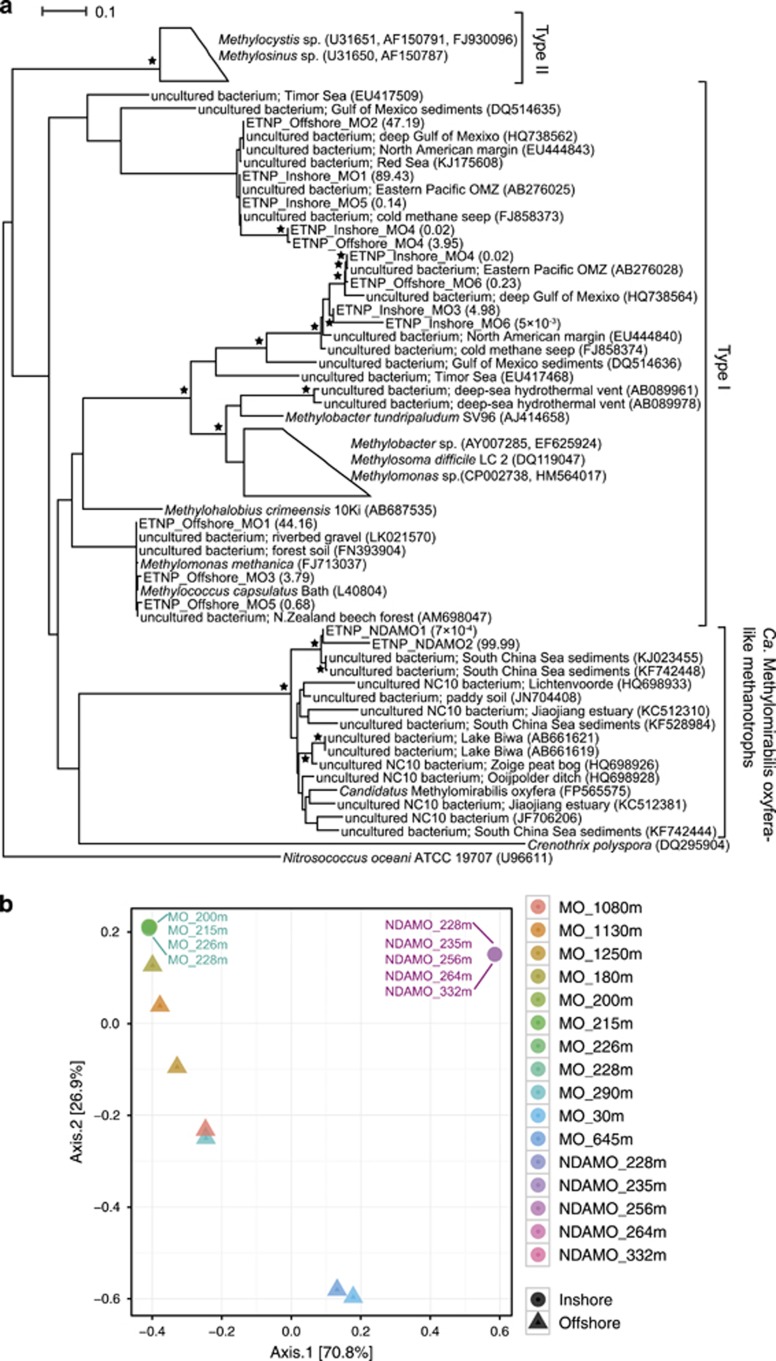
Figure 5.
Aerobic and anaerobic methanotroph community of the water column. (a) Maximum likelihood tree of representative partial pmoA sequences (about 347 bp). Collapsed branches are indicated by a polygon. The amoA gene of Nitrosococcus oceani ATCC 19707 (U96611) was used as the outgroup. Asterisks indicate local support values over 75%. The number in parenthesis following the ETNP_Inshore_MO, ETNP_Offshore_MO and ETNP_NDAMO sequences indicates the % relative abundance of each OTU over the total number of Inshore, Offshore and N-DAMO OTUs, respectively (see Supplementary Table S3 for details). The bar represents 0.1 average nucleotide substitutions per base. (b) Principal coordinate analysis (PCoA) plot of the methanotroph community based on the pmoA sequences and a maximum likelihood tree, and, constructed with the weighted Unifrac metric. Circles indicate inshore samples and triangles indicate offshore samples. MO stands for aerobic methanotrophs and N-DAMO for anaerobic methanotrophs. MO_Inshore and N-DAMO communities are shown by overlapping green and purple circles, respectively.