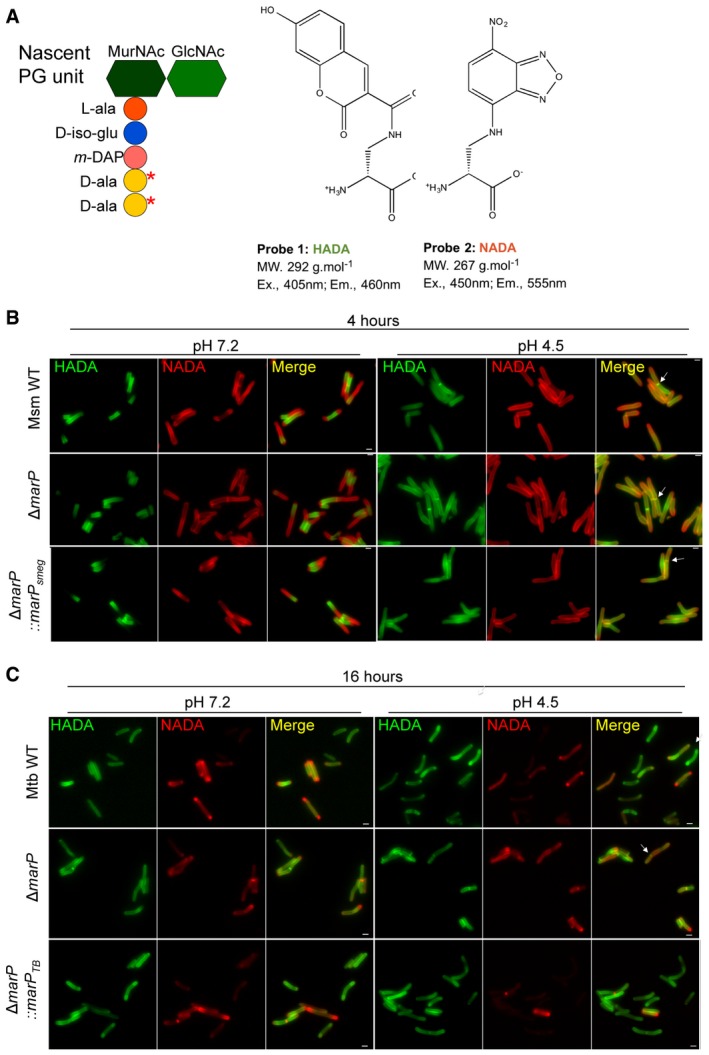
Schematic representing a PG unit and the structure of the fluorescent D‐alanine analogs HADA and NADA. Red stars indicate where the D‐alanine analogs incorporate. MurNAc, N‐acetylmuramic acid; GlcNAc, N‐acetylglucosamine; L‐ala, L‐alanine; D‐iso‐glu, D‐isoglutamic acid; m‐DAP, meso‐diaminopimelic acid; D‐ala, D‐alanine.
Representative images of Msm WT, ΔmarP, and ΔmarP::marP
smeg incubated in 7H9 medium containing 1 mM HADA for 4 h, washed, and further incubated in Sauton's medium at pH 7.2 or at pH 4.5 containing 1 mM NADA for 4 h. Arrows point to septa labeled with both HADA and NADA. Scale bars, 1 μm.
Representative images of Mtb WT, ΔmarP
TB
, and ΔmarP::marP
TB incubated in 7H9 medium containing 1 mM HADA for 24 h, washed, and further incubated in Sauton's medium at pH 7.2 or at pH 4.5 containing 1 mM NADA for 16 h. Arrow points to bacterium with a septum. Scale bars, 1 μm.