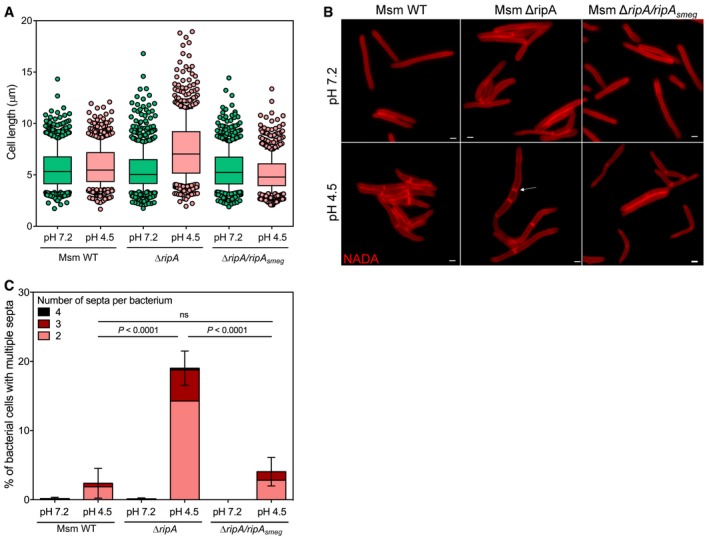
Boxplot of the lengths (μm) of Msm WT, ΔripA, and ΔripA/ripA
smeg incubated for 24 h in Sauton's medium at pH 7.2 or pH 4.5, measured in three independent experiments. Lower and upper whiskers extend to the 10th and 90th percentiles, respectively. The middle bar represents the median and the lower and upper box limits are the 25th and 75th percentiles. The P‐values have been computed using the Ranksum test adjusted for multiple testing. For Msm WT pH 7.2, n = 335, 213, 202; pH 4.5, n = 395, 216, 180. For ΔripA pH 7.2, n = 367, 214, 207; pH 4.5, n = 404, 149, 113. For ΔripA/ripA
smeg pH 7.2, n = 323, 281, 181; pH 4.5, n = 350, 186, 192. n indicates the number of cells in experiments 1, 2, and 3. Adjusted P‐values compared to WT pH 7.2: ΔripA pH 7.2 P = 8.4 × 10−5
; ΔripA/ripA
smeg pH 7.2, P = 6.7 × 10−3. Adjusted P‐values compared to WT pH 4.5: ΔripA pH 4.5, P = 1.3 × 10−15; ΔripA/ripA
smeg pH 4.5, P = 7.7 × 10−4.