Figure 7. Model for the activation of RipA‐mediated PG hydrolysis by MarP in acid‐stressed Mtb.
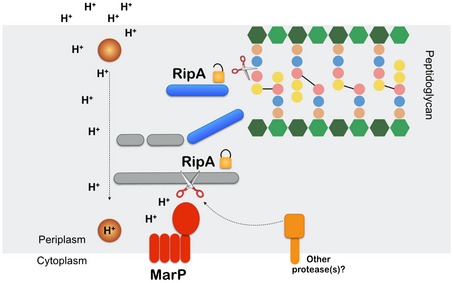
When Mtb faces an acidic environment, the periplasmic protease MarP (red) may sense the stress. Alternatively, MarP might be activated by an unknown protein (brown circle) that senses pH reduction. MarP cleaves the inactive full‐length RipA (gray). RipA processing leads to the release of its C‐terminal fragment that contains a PG hydrolase domain (blue). In other conditions, other proteases (orange) may activate RipA. Ultimately, PG hydrolysis by activated RipA mediates PG remodeling and/or separation of the progeny pair, processes that may sustain Mtb's survival when subjected to acid stress.
