Abstract
Satellite cells, also known as muscle stem cells, are responsible for skeletal muscle growth and repair in mammals. Pax7 and Pax3 transcription factors are established satellite cell markers required for muscle development and regeneration, and there is great interest in identifying additional factors that regulate satellite cell proliferation, differentiation, and/or skeletal muscle regeneration. Due to the powerful regenerative capacity of many zebrafish tissues, even in adults, we are exploring the regenerative potential of adult zebrafish skeletal muscle. Here, we show that adult zebrafish skeletal muscle contains cells similar to mammalian satellite cells. Adult zebrafish satellite-like cells have dense heterochromatin, express Pax7 and Pax3, proliferate in response to injury, and show peak myogenic responses 4–5 days post-injury (dpi). Furthermore, using a pax7a-driven GFP reporter, we present evidence implicating satellite-like cells as a possible source of new muscle. In lieu of central nucleation, which distinguishes regenerating myofibers in mammals, we describe several characteristics that robustly identify newly-forming myofibers from surrounding fibers in injured adult zebrafish muscle. These characteristics include partially overlapping expression in satellite cells and regenerating myofibers of two RNA-binding proteins Rbfox2 and Rbfoxl1, known to regulate embryonic muscle development and function. Finally, by analyzing pax7a; pax7b double mutant zebrafish, we show that Pax7 is required for adult skeletal muscle repair, as it is in the mouse.
Keywords: Pax transcription factors, Rbfox RNA-binding proteins, Muscle stem cells, Myogenesis, Muscle injury
INTRODUCTION
Analysis of frog leg muscles by electron microscopy led to the discovery of mononuclear cells located between the plasmalemma of muscle fibers and the surrounding basement membrane (Mauro, 1961). These cells, termed satellite cells based on their location, were hypothesized to play an important role in muscle regeneration and to function as adult muscle stem cells. Satellite cells have since been established as adult muscle stem cells; they are set aside early in development and remain as a largely quiescent cell population into adulthood (Bentzinger et al., 2012; Montarras et al., 2013). Satellite cells are activated in response to injury and give rise to differentiating myogenic precursor cells (MPCs) (Zammit et al., 2006), as well as other differentiated cell types including bone and brown fat (Asakura et al., 2001; Seale et al., 2008; Shefer et al., 2004).
Mammalian satellite cells reliably express the Pax7 transcription factor (Dumont et al., 2015; Seale et al., 2000). Additionally, Pax3 is expressed in satellite cells in certain mammalian skeletal muscle tissues such as the diaphragm (Montarras et al., 2005). Studies in mouse have identified several transcription factors that regulate satellite cell specification, proliferation, and differentiation (Bentzinger et al., 2012; Davis et al., 1987; Dumont et al., 2015; Rudnicki and Jaenisch, 1995; Weintraub et al., 1991). The most extensively studied of these is Pax7, which is required for murine satellite cell specification, maintenance, and adult skeletal muscle regeneration (Gunther et al., 2013; Seale et al., 2000; von Maltzahn et al., 2013).
Because zebrafish show a high capacity for regeneration, even in some adult tissues for which mammalian counterparts regenerate poorly (Poss et al., 2002; Reimer et al., 2008), the model has emerged as one in which to probe stem cell and regenerative biology. It is well established that larval zebrafish muscle contains satellite-like cells (Devoto et al., 2006; Feng et al., 2006; Gurevich et al., 2016; Knappe et al., 2015; Li et al., 2013; Nord et al., 2013; Otten et al., 2012; Seger et al., 2011; Siegel et al., 2013; Stellabotte et al., 2007; Windner et al., 2015), suggesting that they may also be present in adult muscle. Separate studies have noted the presence of Pax7-positive cells in adult zebrafish skeletal muscle and on isolated myofibers from adult zebrafish (Hollway et al., 2007; Tee et al., 2012; Zhang and Anderson, 2014); however, it is still unclear how similar zebrafish satellite-like cells are to mammalian satellite cells. Definitions of regeneration and repair vary in the literature; for this study, we use the term ‘regeneration’ to indicate replacement of a large portion of a tissue or organ and ‘repair’ to indicate the healing of a damaged portion of a tissue or organ. Zebrafish are competent in both regeneration (e.g., regeneration of the heart; Poss et al., 2002) and repair (e.g., healing after focal needle-stick injury; Gurevich et al., 2016; Knappe et al., 2015; März et al., 2011; Rowlerson et al., 1997; Seger et al., 2011). A few studies have examined skeletal muscle repair (Rowlerson et al., 1997; Tee et al., 2012) and regeneration (Saera-Vila et al., 2015; Saera-Vila et al., 2016) after injury in adult zebrafish. The work from Rowlerson and colleagues (1997) was performed before the identification of satellite cell markers such as Pax7 and Pax3, and thus did not address whether adult zebrafish possess myogenic progenitor cells that function as satellite cells. A second study (Tee et al., 2012) observed Pax7-positive cells after needle-stick injury of adult zebrafish trunk musculature; however, only one time-point was examined and the contribution of Pax7-expressing cells to muscle repair was not assessed. Most recently, regeneration of extraocular muscle was found to occur through Fgf-regulated myocyte dedifferentiation that does not employ classic Pax7-positive satellite-like cells (Saera-Vila et al., 2015; Saera-Vila et al., 2016). Here, we have undertaken a comprehensive study to define the cells employed and steps involved during trunk skeletal muscle repair.
In addition to known satellite cell markers Pax7 and Pax3, there is active interest in identifying additional markers that may mark and/or regulate satellite cells and regenerating skeletal muscle. We have shown previously that Rbfox1l and Rbfox2, members of the Rbfox family of RNA binding proteins, are required for zebrafish embryonic muscle development and function (Gallagher et al., 2011). In mammals, the Rbfox family includes Rbfox1 (A2BP1; HRNBP1; FOX-1), Rbfox2 (RBM9; FOX-2) and the neural-specific Rbfox3 (NeuN; FOX-3) (Kuroyanagi et al., 2009). Rbfox proteins regulate alternative splicing in metazoans and their target intronic binding sites are extensively conserved among vertebrates (Gallagher et al., 2011; Lovci et al., 2013; Minovitsky et al., 2005; Yeo et al., 2007). Human RBFOX2 is required for human embryonic stem cell survival (Yeo et al., 2009) and promotes differentiation of pluripotent stem cells (Venables et al., 2013), suggesting a potential role for RBFOX2 in stem or progenitor cell regulation in multiple tissue types and organ systems. During zebrafish embryonic myogenesis, rbfox2 is expressed in the presomitic mesoderm (a muscle progenitor domain) and newly-formed myotomal segments, but only weakly as muscle progenitors differentiate, whereas rbfox1l is expressed weakly in the progenitor domain and strongly in differentiating muscle cells (Gallagher et al., 2011). Previous studies corroborate muscle-specific embryonic expression of rbfox1l (Baxendale et al., 2009; Jin et al., 2003). Given these early expression patterns, we hypothesized that Rbfox2 and Rbfox1l may mark satellite-like cells and newly-forming myofibers, respectively, during injury-induced skeletal muscle repair in adult zebrafish.
In this study, we investigated the process of muscle repair in adult zebrafish skeletal muscle. Using transmission electron microscopy (TEM), immunohistochemistry, and transgenic reporter lines, we identify cells that closely resemble satellite cells within adult zebrafish skeletal muscle. Mechanical injury results in robust activation of Pax7 and pax3a:GFP-expressing satellite-like cells by 4–5 dpi. Acute EdU labeling of S-phase cells indicates that adult zebrafish satellite-like cells have proliferative potential, and perdurance of pax7a-driven GFP in new myofibers that form after muscle injury suggests that new muscle is at least partially derived from satellite-like cells. Lastly, our analysis of adult pax7a; pax7b double mutant zebrafish reveals that Pax7 function is required for adult skeletal muscle repair. Our findings further establish the adult zebrafish as a model to study satellite cell biology, and set the stage for future studies assessing Rbfox1l and Rbfox2 function in satellite cells, injury-induced repair, and muscle disease.
RESULTS
Pax7-expressing satellite-like cells are found within adult zebrafish skeletal muscle
Satellite cells were originally identified by electron microscopy (EM) in frog leg muscle as cells with dense heterochromatin (Mauro, 1961). Transmission electron microscopy (TEM) studies have also identified satellite cells in mammals (Seale et al., 2000). We performed TEM to determine whether cells resembling satellite cells are present in adult zebrafish skeletal muscle and find cells containing dense nuclear heterochromatin in both slow- and fast-twitch muscle fiber domains (Fig.1A, B; white arrowheads) that are easily distinguished from muscle fiber nuclei (myonuclei) (Fig. 1A, B; blue arrowheads). As in mammals (Seale et al., 2000), these satellite-like cells are located outside of the muscle fiber membrane and are surrounded by basal lamina (Fig. 1A′, B′; red arrows). In addition to morphological criteria, mammalian satellite cells also reliably express the Pax7 transcription factor (Seale et al., 2000). Previous studies have shown that Pax7-positive cells are present in larval zebrafish and are activated in response to injury and under disease conditions (Berger et al., 2010; Knappe et al., 2015; Seger et al., 2011). As described previously (Hollway et al., 2007; Tee et al., 2012), we also find Pax7-positive cells in adult zebrafish muscle (Fig. 1C, D). Pax7-positive cells are located beneath the basal lamina of the surrounding basement membrane (Fig. 1C–C‴) and external to the muscle membrane, which is marked in a Dystrophin FlipTrap insertion line, dmdct90aGt, that expresses a functional Dystrophin-Citrine fusion protein (Trinh et al., 2011) (Fig. 1D, D′). These data show that satellite-like cells in adult zebrafish express well-described satellite cell markers and are located in positions consistent with the location of satellite cells in other organisms (Bentzinger et al., 2012).
Figure 1. Cells with satellite cell features are present in adult zebrafish skeletal muscle.
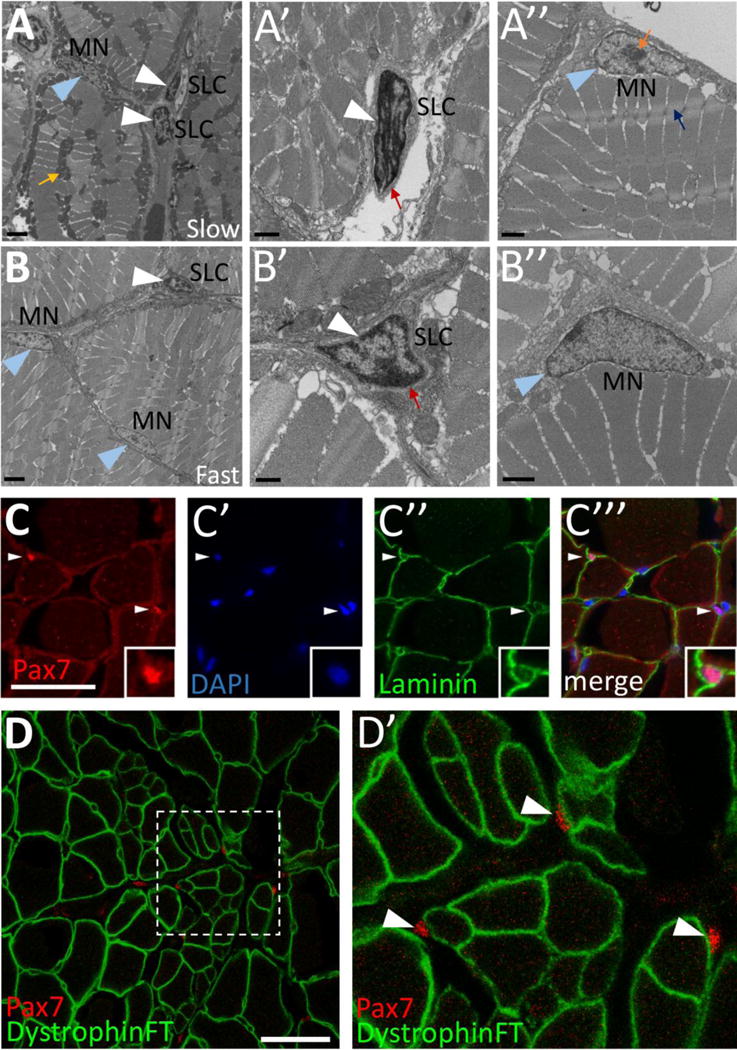
(A–B) Transmission electron microscopy (TEM) on adult zebrafish tail skeletal muscle from slow-twitch (A–A″) and fast-twitch (B–B″) muscle fiber domains. (A) TEM of slow muscle reveals mitochondrial-rich muscle fibers (mustard arrow points to a cluster of dark gray mitochondria), putative satellite-like cells (SLCs; white arrowheads), and myonuclei (MN; blue arrowhead points to a myonucleus). (A′) Higher magnification view of a SLC containing dense heterochromatin (white arrowhead) and surrounded by basal lamina (red arrow). (A″) A myonucleus (MN; blue arrowhead) located within the muscle fiber membrane. Myonuclei are characterized by a lack of dense heterochromatin and often by a prominent nucleolus (orange arrow). An adjacent myofibril is indicated by a blue arrow. (B) TEM of fast muscle reveals fewer mitochondria, a putative SLC (white arrowhead), as well as myonuclei (MN, blue arrowheads). (B′) Higher magnification view of a SLC containing dense heterochromatin (white arrowhead) and surrounded by basal lamina (red arrow). (B″) A myonucleus (MN; blue arrowhead) located within the muscle fiber membrane shows similar characteristics to slow muscle myonuclei. (CC′″) Pax7-positive cells (red; white arrowheads) are localized within the basal lamina of adult zebrafish skeletal muscle (anti-Laminin in green) (inset shows enlarged view of Pax7-positive cell). DAPI labeling confirms Pax7 localization in nuclei. (D–D′) Pax7-positive cells (red; arrowheads) are located outside of the muscle fiber membrane, marked by a Dystrophin FlipTrap transgenic line. (D′) Magnified view of boxed region in D. Scale bar in A, B is 2 μm, in A′, A″, B″ is 1 μm, in B′ is 500 nm, and in C and D is 30 μm.
To evaluate the pax7a:GFPi131 BAC transgenic zebrafish line (Seger et al., 2011) as a marker for adult satellite-like cells, we examined overlap of Pax7 and pax7a:GFP expression in uninjured muscle sections (Fig. S1A–B″). As expected, almost all Pax7-positive cells express the pax7a:GFP transgene (98%; 302/308 cells; n=3). The converse comparison reveals that although the majority of pax7a:GFP transgene-expressing cells express Pax7 (72%; 302/422 cells; n=3), some are Pax7-negative (28%; 120/422 cells; n=3), possibly reflecting GFP perdurance, ectopic transgene expression, or cytoplasmic cellular protrusions for which the nucleus is out of the plane of section. To further clarify, we performed experiments to assess overlap of pax7a:GFP, Pax7, and DAPI labeling (Fig. S1C–D‴). Only 11% of pax7a:GFP-positive cells lack a DAPI-positive nucleus (42/398 cells; n=3) and thus likely represent cytoplasmic protrusions of cells for which nuclear expression cannot be assessed. Of the remaining pax7a:GFP-positive, DAPI-positive cells, the majority are Pax7-positive (70%; 248/356 cells; n=3); those that are Pax7-negative (30%; 108/356 cells; n=3) likely reflect GFP perdurance or ectopic transgene expression. Finally, virtually all Pax7-positive cells are DAPI-positive (253/255 cells; n=3) (Fig. 1C–C‴), further validating Pax7 as a bona fide satellite-like cell nuclear marker.
We find that the pax3a:GFPi150 BAC transgene (Seger et al., 2011) is also expressed in adult zebrafish satellite-like cells (Fig. S2). In uninjured adult muscle, a large fraction of Pax7-expressing cells express the pax3a:GFP transgene (90%; 75/84 cells; n=3), and conversely, a majority of pax3a:GFP-expressing cells also express Pax7 (67%; 75/112 cells; n=3) (Fig. S2C, C′, C‴, mustard arrowheads). Although we utilize pax3a:GFP and pax7a:GFP transgenes as satellite-like cell markers in this work, we note that both are also expressed in non-myogenic cells in the skin (pax3a:GFP expression at the outer edge in Fig. S2A; pax7a:GFP expression at the outer edge in Fig. S2B) and spinal cord (Fig. S2A, arrowhead, for pax3a:GFP expression; Fig. S1D, for pax7a:GFP expression). In subsequent experiments, a nuclear label (Pax7, DAPI, or EdU) is usually included to assess and quantify marker and/or transgene expression.
Satellite-like cells are localized predominantly in the slow muscle domain
As in mammals, zebrafish slow muscle fibers contain many mitochondria compared to fast muscle fibers (Fig. 1A, B; an example indicated by a mustard arrow) (Schiaffino and Reggiani, 2011; Talbot and Maves, 2016; van Raamsdonk et al., 1980; Waterman, 1969), consistent with the distinct oxidative capacity of these two cell types. However, unlike in mammals, where slow and fast fiber types are often intermingled within muscles, zebrafish slow and fast fibers are segregated into spatially distinct domains (see Fig. 2A) (Bassett and Currie, 2003; Devoto et al., 1996; Elworthy et al., 2008; Jackson et al., 2015; Ju et al., 2003; Nord et al., 2014; Waterman, 1969). To identify slow and fast muscle domains, we used two transgenic lines: Tg(smyhc1:GFP)i104 that expresses GFP in slow muscle (Fig. 2C, D) (Elworthy et al., 2008) and Tg(mylpfa:mCherry)cz3327 that expresses mCherry in fast muscle (Fig. 2E, F) (Ignatius et al., 2012). These two transgenes robustly identify the majority of muscle fibers in the respective slow and fast muscle domains, but there are some non- or weakly-expressing cells at the border between them that may be intermediate fiber types (Elworthy et al., 2008; van Raamsdonk et al., 1980; Waterman, 1969). As expected, we find that satellite-like cells are sparsely localized throughout most of the adult myotome; however, they are enriched near the horizontal myoseptum, a slow muscle domain (schematized in Fig. 2A). To quantify the difference, we counted Pax7-positive cells within domains definitively labeled as slow muscle (Fig. 2C, D; cells to the left of the dotted blue line in the symhc1:GFP-positive domain) or fast muscle (Fig. 2E; cells to the right of the dotted brown line in the mylpfa:mCherry-positive domain) and normalized to total myofiber number in each domain. We find that Pax7-positive cells are 21-fold enriched in slow muscle relative to fast muscle (Fig. 2B), with very few Pax7-positive cells seen deep within the ventral myotome, a fast muscle domain (Fig. 2F).
Figure 2. Adult zebrafish satellite-like cells are concentrated predominantly in slow muscle.
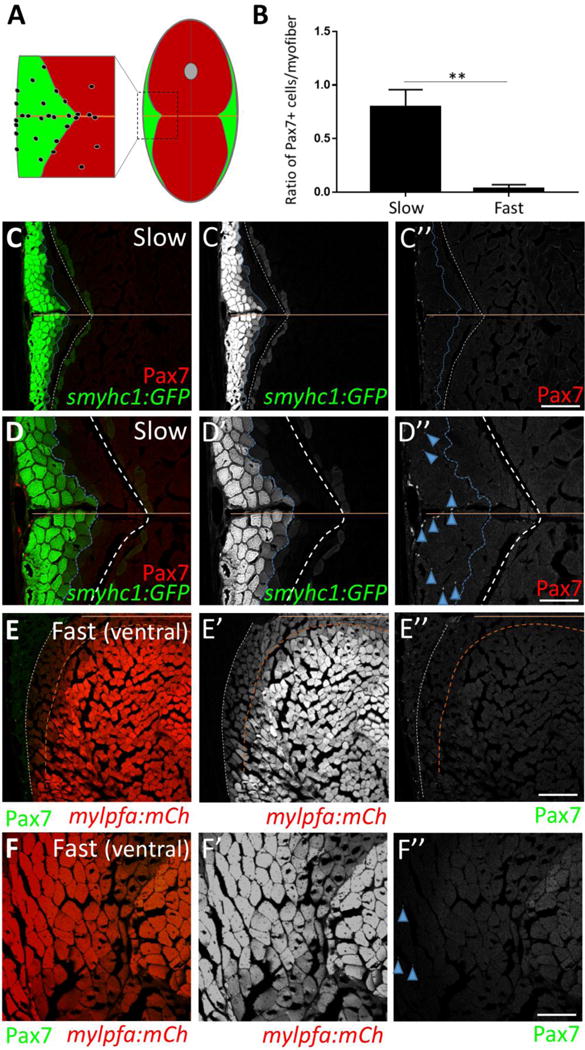
(A) Schematic of cross-section through adult zebrafish trunk musculature, depicting fast-twitch (red) and slow-twitch (green) muscle fiber domains and a concentration of SLCs (black dots) within the slow muscle domain. The horizontal myoseptum is indicated by a brown line; the gray circle at the dorsal midline depicts the spinal cord. (B) The number of Pax7-positive cells, normalized to total myofiber number, is significantly higher in slow versus fast muscle; Student’s t-test (p**<0.01). (C–C″) Pax7-positive cells (in red) are more common within slow muscle. Slow muscle fibers are marked with smyhc1-driven GFP (left of the dotted blue line). The boundary between slow and fast muscle fiber domains is marked by a dotted white line. The area between dotted blue and dotted white lines may represent muscle fibers with an intermediate identity. The horizontal myoseptum is indicated by a brown line. (D–D″) Higher magnification view of slow muscle. For quantification shown in B, Pax7-positive cells (D″; examples depicted by blue arrowheads) within the smyhc1:GFP-positive domain (left of the dotted blue line) were counted and normalized to the number of smyhc1:GFP-positive myofibers in the same area. (E–E″) Pax7-positive cells (in green) are rare within ventral fast muscle. Fast (ventral) muscle fibers are marked with mylpfa-driven mCherry (right of the dotted brown line). The area between dotted brown and white lines (weakly expressing mylpfa:mCherry) may represent intermediate muscle fiber identity. A dotted white line separates fast and slow muscle domains. (F–F″) Higher magnification image of fast muscle. For quantification shown in B, Pax7-positive cells (F″; blue arrowheads) within the mylpfa:mCherry-positive domain were counted and normalized to the number of mylpfa:mCherry-positive myofibers in the same area. Scale bar in C” and E” is 150 μm and in D” and F” is 75 μm.
Mechanical injury of adult zebrafish skeletal muscle results in Pax7-positive satellite-like cell proliferation and a concomitant increase in Pax7-positive cells at the injury site
Pax7-expressing satellite cells are required for skeletal muscle regeneration after injury in mammals (Lepper et al., 2011; Sambasivan et al., 2011). To determine whether adult zebrafish satellite-like cells behave similarly, we injured tail skeletal muscle by a single needle stick as previously described (Rowlerson et al., 1997) in the ventral myotome, to avoid the dorsally-located spinal cord and the satellite-like cell populations near the horizontal myoseptum (Fig. 3A). Fish were sacrificed daily from 2–7 dpi to assess whether and when satellite-like cells are activated by skeletal muscle injury (Fig. 3A). We find that Pax7-positive satellite-like cells increase after injury, peaking at 4 dpi and declining thereafter (Fig. 3B; Fig. S3G). The increase in Pax7-positive cells in the injured region contrasts sharply to the minimal number of Pax7-positive cells found within the same ventral myotome region of uninjured fish (see Fig. 2B, F″).
Figure 3. Satellite-like cells in adult zebrafish skeletal muscle proliferate after mechanical injury.
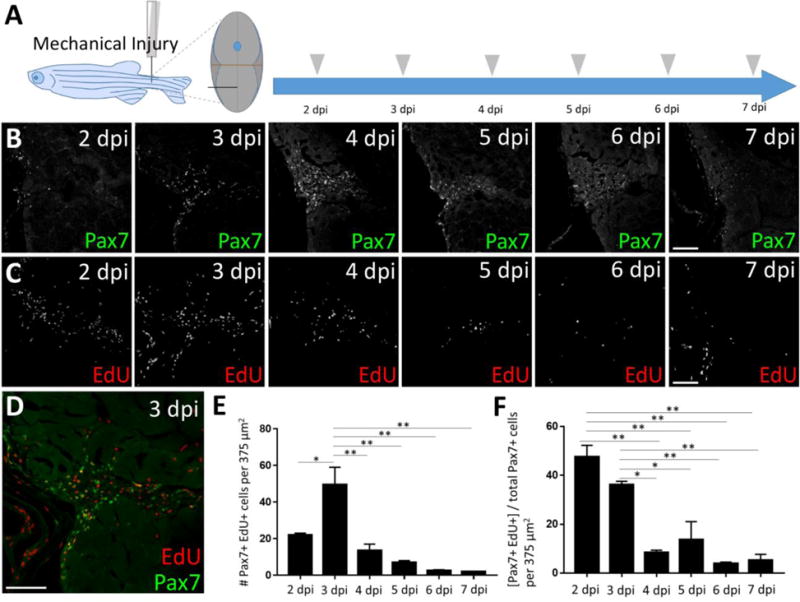
(A) Schematic of experimental procedure, in which mechanical injury is performed by a single needle-stick into tail skeletal muscle. The approximate position of needle-stick injury in the ventral myotome is indicated in cross-sectional view. EdU was administered (gray arrowhead) by intraperitoneal injection at each post-injury time-point, allowing for EdU incorporation into S-phase cells, and fish were sacrificed 4 hours later (acute EdU labeling). Tissue was collected at 2, 3, 4, 5, 6, and 7 dpi. (B) Pax7-positive cell numbers increase at the injury site and peak at 4 days post-injury (dpi). (C) Acute EdU labeling indicates robust proliferation at the injury site at 2 and 3 dpi that declines thereafter. (D) Overlap of Pax7 and EdU at 3 dpi indicates satellite-like cell proliferation post-injury. (E) Quantification of Pax7 and EdU double positive cells at each post-injury time-point reveals a peak number of proliferating satellite-like cells at 3 dpi in an 375 μm2 area encompassing the injury site; p*<0.05; p**<0.01. (F) Quantification of the percentage of Pax7-positive cells that are EdU-positive (out of total Pax7-positive cell population) at each post-injury time-point indicates that nearly 50% of Pax7-positive cells are proliferating at 2 dpi, and ~35% are proliferating at 3 dpi. The percentage of Pax7-positive cells that are EdU-positive at later post-injury time-points (4–7 dpi) is significantly less; p*<0.05; p**<0.01. One-way ANOVA followed by Tukey’s multiple comparisons test was performed for statistical analyses. Scale bar in all panels is 75 μm.
To assess proliferation after injury, we acutely labeled cells using EdU at each post-injury time-point and observed the greatest numbers of EdU-positive cells at 2 and 3 dpi (Fig. 3C; Fig. S3A–F, H). Consistent with the expectation that non-activated satellite cells are a quiescent, non-dividing population, few Pax7-positive cells show EdU label in uninjured fish (Fig. S2C′, C″, C‴, white arrowheads); by contrast, many Pax7-positive, EdU-positive cells are detected after injury, peaking at 3 dpi (Fig. 3D, E; Fig. S3A–F). Additionally, the percentage of Pax7-positive cells that are EdU-positive is nearly 50% at 2 dpi, ~35% at 3 dpi, and less than 10% by 4 dpi (Fig. 3F). Between 2 and 4 dpi, we observe a 3-fold increase in the number of Pax7-positive cells at the injury site (Fig. S3G), suggesting an expansion of the Pax7-expressing cell population as a result of proliferation during that timeframe. Taken together, these data show that Pax7-expressing satellite-like cells in adult zebrafish skeletal muscle proliferate locally and robustly after needle stick injury.
Most Pax7-positive satellite-like cells express pax3a:GFP during adult zebrafish skeletal muscle repair
After needle stick injury in the pax3a:GFPi150 transgenic line (Seger et al., 2011), we observe that pax3a:GFP-positive cell number, like Pax7-positive cell number, gradually increases at the injury site and peaks in parallel with Pax7-positive cells (Fig. S4A, B). By 5 dpi, we observe a localized and robust activation of pax3a:GFP expression at the injury site (Fig. S4D). Satellite-like cells that are Pax7-positive and pax3a:GFP-positive are found at the injury site during a time of peak satellite-like cell response (Fig. S4D, D′, white arrowheads; Fig. S5D, G, white arrowheads). Quantification at 4 dpi indicates that 94% of Pax7-positive cells are also pax3a:GFP-positive (Fig. S5G, white arrowheads, 544/575 total cells counted; n=3), whereas only 6% are pax3a:GFP-negative (Fig. S4D′, blue arrowheads; Fig. S5G, blue arrowheads, 31/575 total cells counted; n=3). Due to the cytoplasmic localization of GFP, it is difficult to accurately quantify the percentage of pax3a:GFP-positive cells that express Pax7 in sections (pax3a:GFP-positive, Pax7-negative cells are shown by purple arrowheads in Fig. S4D′ and Fig. S5G). Since a large fraction of Pax7-expressing cells in uninjured muscle also express pax3a:GFP, it is not surprising that we find that cells expressing both Pax7 and pax3a:GFP proliferate in response to injury (Fig. S5C, mustard arrowheads; Fig. S5D–G, mustard arrowheads in G; compare with proliferation of Pax7-positive cells in Figure 3). These data show that satellite-like cells expressing both Pax7 and pax3a:GFP increase in number near the injury site after skeletal muscle needle stick injury.
Myoblast- and/or myogenic precursor-like cells that express myf5:GFP and myoD:GFP are present during adult zebrafish skeletal muscle repair
At 4 dpi, we observe cells that are Pax7-negative, pax3a:GFP-positive, and EdU-positive at the injury site (Fig. S5C, brown arrowheads). We hypothesized that these cells may be derived from satellite-like cells that have down-regulated Pax7 (and are likely no longer actively transcribing pax3a:GFP) and thus represent proliferating myogenic precursor cell (MPC)-like cells or myoblasts that are progressing toward differentiation. In mammals, MPCs and myoblasts express the myogenic regulatory factors Myod and Myf5 (Bentzinger et al., 2012), while late-stage myoblasts express Myogenin (Myog) as they differentiate into myofibers. To determine whether myoblasts are present after injury, we performed injury in the myod:GFPi124 BAC transgenic line (Seger et al., 2011). We find myod:GFP-positive cells at the injury site at 4 dpi, some of which label with an acute pulse of EdU, suggesting that injury induces a proliferating myoblast-like population in adult zebrafish (Fig. 4A–A‴, arrowheads). myod:GFP-positive cells make up 15%, on average, of the total proliferative population present at the injury site (4 dpi), slightly fewer than (but not significantly different from) the percentage of proliferating Pax7-positive cells present (25%; Fig. 4B–C). The number of cells expressing both myod:GFP and Pax7 is minor; the large majority of Pax7-positive cells are myod:GFP-negative (90%; Fig. 4B–B‴, white arrowheads; Fig. 4D), further suggesting the existence of a myogenic population of cells distinct from (or possibly derived from) the Pax7-expressing satellite-like cell population. Additionally, we further investigated the presence of myoblast-like cells by performing injury in adult zebrafish carrying two transgenes: -80.0myf5:GFPzf37 (myf5-driven GFP, hereafter ‘myf5:GFP’; Chen et al., 2007) and myog:H2B-mRFP (myog-driven H2B (nuclear)-mRFP; Tang et al., 2016) (Fig. 4E). At 5 dpi, we find myf5:GFP-positive cells at the injury site, some of which are also myog:H2B-mRFP-positive (Fig. 4E–E″). The former are presumptive myoblasts or MPCs, whereas the latter may be late-stage myoblasts or newly-forming myofibers in which myf5-driven GFP perdures (Fig. 4E″, arrowheads). These data establish the presence of a proliferating myoblast-like population post-injury in adult zebrafish skeletal muscle.
Figure 4. Myoblast-like cells, detected using myf5:GFP and myod:GFP transgene expression, are present post-injury in adult zebrafish skeletal muscle.
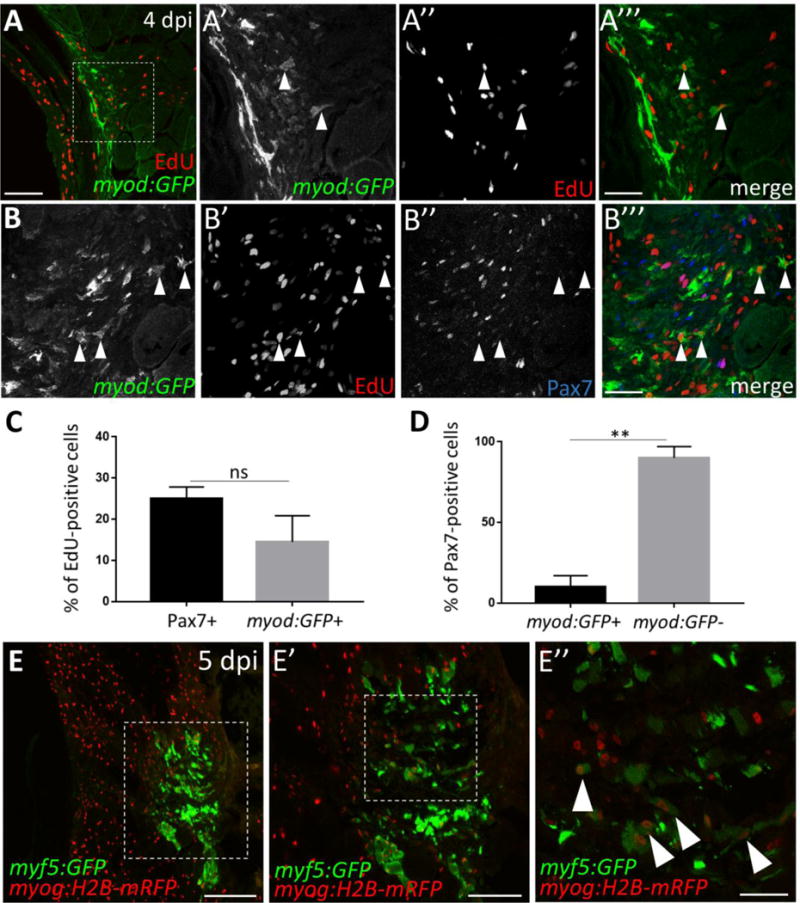
(A) The myod:GFP transgenic line identifies myoblast-like cells at 4 dpi. (A′–A′″) Magnified view of boxed area in A, showing myod:GFP expression (A′), EdU incorporation after an acute EdU pulse (A”), and the overlay (A′″) at 4 dpi. Double-positive cells are indicated by arrowheads. (B–B′″) At 4 dpi, myod:GFP-expressing cells are largely Pax7-negative (examples indicated by arrowheads). (C) Percentage of total proliferating cells (i.e. those that incorporate EdU) at 4 dpi that are Pax7-positive (25%) versus myod:GFP-positive (15%). The percentage of proliferating cells that are Pax7-positive (25%) is similar to that shown in Fig. S3I and is slightly higher than the percentage of proliferating cells that are myod:GFP-positive (15%), although the difference is not statistically significant. (D) Percentage of Pax7-expressing cells at 4 dpi that are myod:GFP-positive (10%) versus myod:GFP-negative (90%), p**<0.01. (E) myf5:GFP-positive cells are present at the injury site at 5 dpi. (E′) Magnified view of boxed region in E. (E”) Magnified view of boxed region in E′ reveals myf5:GFP-positive cells that are myog:H2B-mRFP-positive (arrowheads). Scale bar in A and E′ is 75 μm, in E is 150 μm, and in A′–B′″ and E” is 30 μm.
Rbfox2 and Rbfox1l RNA-binding proteins are differentially expressed in Pax7-positive satellite-like cells during adult muscle repair
We previously showed that zebrafish rbfox1l and rbfox2 genes are critical for embryonic skeletal muscle development and function (Gallagher et al., 2011) and hypothesized that they may also play important roles in adult skeletal muscle satellite-like cells. We generated antibodies against Rbfox1l and Rbfox2, and verified specificity by western blot of wild-type and Rbfox-depleted embryo extracts (Fig. S6A). In uninjured adult muscle, we find that Rbfox2 is expressed prominently in Pax7-positive cells (Fig. S6B–B″; white arrowheads) and in a minor subset of other nuclei (Fig. S6B–B″; gray arrowheads). Interestingly, some cells within the myotome have nuclear-localized Rbfox2, while in other cells Rbfox2 is cytoplasmic (Fig. S6E). Since Rbfox2 localizes with Pax7 in the nucleus of satellite-like cells (Fig. S6B–B″) we infer that cytoplasmic Rbfox2 marks other, non-myogenic cell types (such as blood or immune cells). In contrast, Rbfox1l is expressed in a subset of Pax7-positive cells (Fig. S6C–C″; white arrowheads) but more prominently in most, if not all, myog:H2B-mRFP-positive adult myofibers (Fig. S6D–D″), consistent with known muscle-specific rbfox1l expression (Baxendale et al., 2009; Gallagher et al., 2011). In uninjured muscle, Rbfox1l is consistently localized to the nucleus, seen by strong overlap with DAPI (Fig. S6F).
To assess Rbfox expression during muscle repair, we performed immunohistochemistry to detect Rbfox1l and Pax7 or Rbfox2 and Pax7 on adjacent sections at post-injury time-points 2–7 dpi (Fig. 5A). Rbfox1l is robustly expressed at the height of the satellite-like cell response (4 dpi; Fig. 5B). The absolute number of Pax7-positive, Rbfox1l-positive cells at the injury site ranges from 10–35 cells, with no significant differences observed among post-injury time-points (Fig. 5B; magnified 4 dpi example shown in Fig. 5C–C″; purple arrowheads). Between 2–7 dpi, the percentage of Pax7-positive cells that are Rbfox1l-positive ranges between 28–43%, with no significant difference observed among post-injury time-points (Fig. 5D). Rbfox2 expression also peaks at 4 dpi, and precedes Rbfox1l expression as deduced by examining adjacent sections (Fig. 5B, E; Fig. 5F–F″; purple arrowheads). The absolute number of Pax7-positive, Rbfox2-positive cells at the injury site ranges from 34–101 cells, with the greatest number observed at 3 and 4 dpi (Fig. 5E), consistent with Pax7-positive cell response post-injury. In contrast with Rbfox1l, post-injury Rbfox2 expression closely mirrors that of Pax7; almost all (98–99.5%) Pax7-positive cells are Rbfox2-positive between 2–7 dpi, with no significant difference observed among time-points (Fig. 5G). We also observe Rbfox2-positive cells that are Pax7-negative (Fig. 5F–F″, blue arrowheads) and thus not satellite cells. These may represent differentiating myogenic cells, or even non-myogenic and/or immune or blood cells, as Rbfox2 is expressed quite broadly in zebrafish embryos (Gallagher et al, 2011) and in lymphocytic B-cells and hematopoietic progenitor cells in mammals (Ponthier et al., 2006). These data show that although RNA-binding proteins Rbfox1l and Rbfox2 are both expressed in quiescent and activated satellite-like cells at some level, Rbfox2 is more prevalent in satellite-like cells than Rbfox1l under both conditions. Additionally, during skeletal muscle repair, Rbfox2 expression precedes Rbfox1l expression at the injury site.
Figure 5. Satellite-like cells express Rbfox1l and Rbfox2 RNA-binding proteins after mechanical injury of adult zebrafish skeletal muscle.
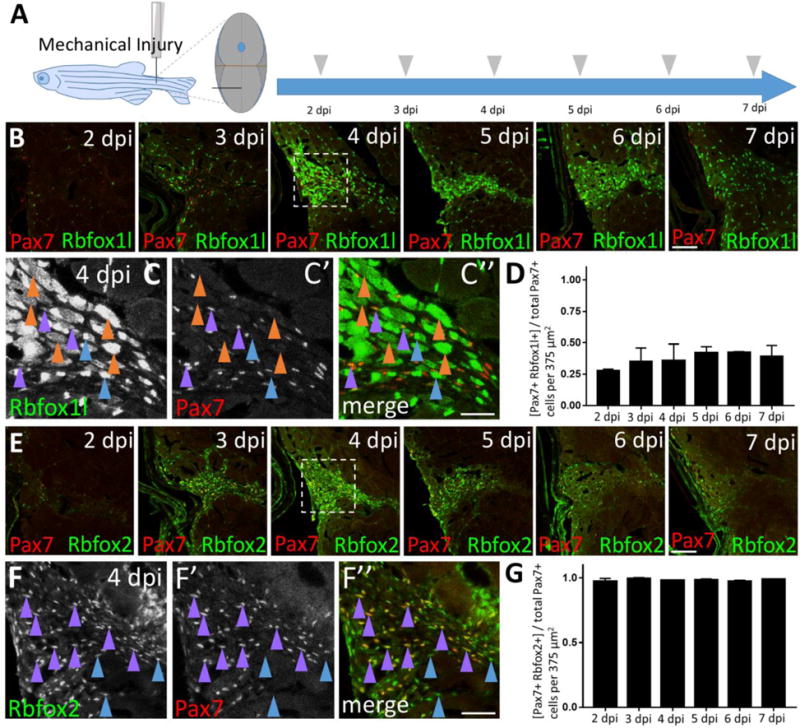
(A) Schematic of experimental procedure, which is identical to that described Fig. 3A. (B) Rbfox1l (green) is robustly expressed beginning at 4 dpi, coinciding with the onset of new myofiber formation. Pax7 expression (overlaid in red) is the same as shown in Fig. 3C; the merge here shows overlap of Pax7 and Rbfox1l expression. The boxed region at 4 dpi is magnified in C–C”. (C–C”) Magnified view of boxed area in B, showing expression of Rbfox1l (C) and Pax7 (C′), and the overlay (C”) at 4 dpi. Examples of Pax7-positive, Rbfox1l-positive cells are indicated by purple arrowheads and of Pax7-negative, Rbfox1l-positive cells by blue arrowheads. Rbfox1l expression is observed in the cytoplasm of presumptive newly-forming myofibers (orange arrowheads indicate examples). Rbfox1l expression in uninjured areas surrounding the injury site appears less uniform in muscle fiber nuclei (compared to Fig. S6 D–D″) as laser intensity was reduced to prevent over-saturation of signals within the injury site. (D) Ratio of Pax7-positive cells that are Rbfox1l-positive at post-injury time-points 2–7 dpi. (E) Onset of Rbfox2 expression (green) at 3 dpi is earlier than Rbfox1l, and closely resembles post-injury Pax7 expression (overlaid in red). The sections are alternating with those in B. The boxed region at 4 dpi is magnified in F–F”. (F–F”) Magnified view of boxed area in C, showing expression of Rbfox2 (F) and Pax7 (F′), and the overlay (F”) at 4 dpi. Examples of Pax7-positive, Rbfox2-positive cells are indicated by purple arrowheads, and of Pax7-negative, Rbfox2-positive cells by blue arrowheads. (G) Ratio of Pax7-positive cells that are Rbfox2-positive at post-injury time-points 2–7 dpi. Scale bar in B and E is 75 μm, and in C–C” and F–F” is 30 μm.
Rbfox1l, in contrast to Rbfox2, is expressed in differentiating muscle cells after injury
We hypothesized that Rbfox-expressing non-satellite cell populations might represent differentiating myogenic cells. To test this, we analyzed Rbfox1l expression post-injury in the myog:H2B-mRFP transgenic line (Fig. 6; myonuclei labeled in red). We find myog:H2B-mRFP-expressing cells at the injury site starting largely at 4 dpi and persisting thereafter, corresponding with onset of new myofiber formation (Fig. 6B). The absolute number of Rbfox1l; myog:H2B-mRFP double-positive cells similarly increases at the injury site (Fig. 6C, D). As in uninjured muscle (Fig. S6D″), Rbfox1l expression post-injury is found mainly within differentiating cells, with myog:H2B-mRFP-positive nuclei accounting for 71–95% of the total Rbfox1l-positive population between 4 and 7 dpi (Fig. 6E, 6F′–H′; white arrowheads). Interestingly, Rbfox1l is expressed in nuclei (Fig. 6F′–H′; white arrowheads) and cytoplasm (Fig. 6F′–H′; brown arrowheads) of newly-forming myofibers.
Figure 6. Rbfox1l is expressed predominantly in the nucleus and cytoplasm of newly-forming myofibers during skeletal muscle repair.
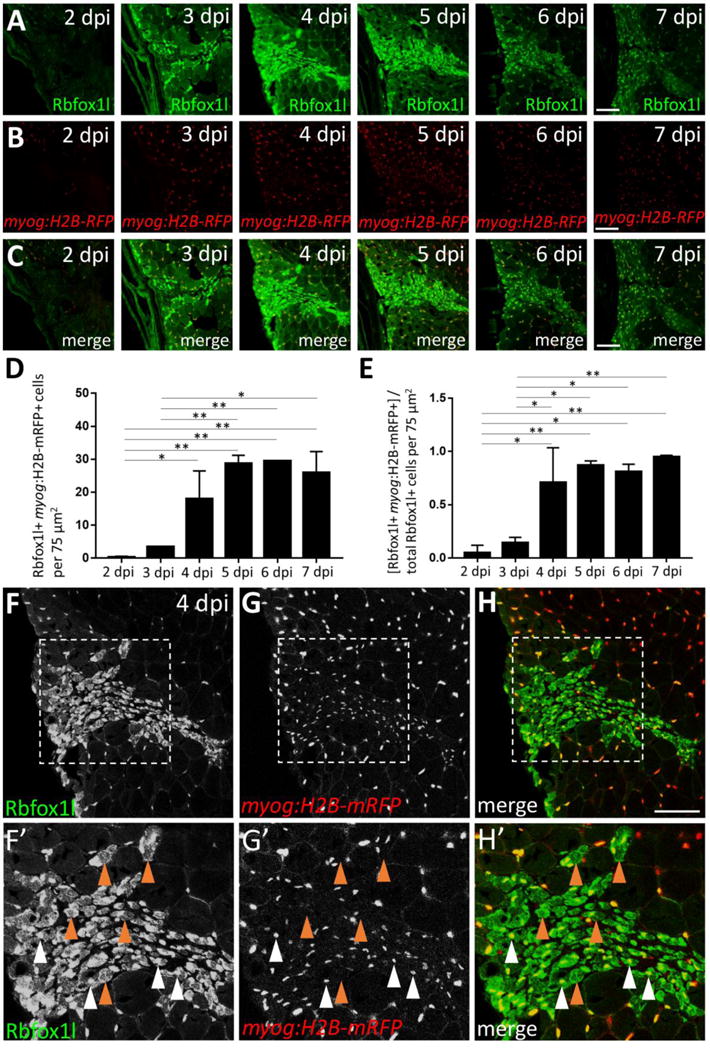
(A) Injury time-course experiment performed in the myog:H2B-mRFP transgenic line showing Rbfox1l expression at each post-injury time-point. (B) myog:H2B-mRFP expression is readily observed within the injury site by 4 dpi. (C) Overlay of Rbfox1l and myog:H2B-mRFP expression. (D) Total number of Rbfox1l, myog:H2B-mRFP double-positive cells at 2–7 dpi in a 75 μm2 area within the injury site (to exclude Rbfox1l-positive; myog:H2B-mRFP-positive myonuclei within surrounding uninjured fibers). (E) Ratio of Rbfox1l-positive cells that are myog:H2B-mRFP-positive at 2–7 dpi. One-way ANOVA followed by Tukey’s multiple comparisons test was performed for statistical analyses in D and E (p*<0.05; p**<0.01). (F–H) Higher magnification view of 4 dpi injury site shown in A–C. (F′–H′) Area shown is the boxed region in F–H. A majority of Rbfox1l-positive nuclei are also positive for myog:H2B-mRFP (examples indicated by white arrowheads), as expected from the large overlap of these two markers in differentiated muscle outside of the injury site. Rbfox1l expression in uninjured areas surrounding the injury site appears less uniform in muscle fiber nuclei in this figure (compared to Fig. S6 D–D″) as laser intensity was reduced to prevent over-saturation of signals within the injury site. Some newly-forming myofibers that express Rbfox1l in the nucleus and/or cytoplasm are myog:H2B-mRFP-negative (examples indicated by brown arrowheads). Scale bar in all panels is 75 μm.
In similar experiments designed to characterize the Rbfox2-expressing non-satellite cell population, we observe that Rbfox2 is expressed in fewer myonuclei during skeletal muscle repair (Fig. 7A–C; F–H′, white arrowheads). Although the number of Rbfox2; myog:H2B-mRFP double-positive cells trends upward after 3 dpi and until 6 dpi, the data are not statistically significant among time-points (Fig. 7D). Additionally, myog:H2B-mRFP-positive myonuclei only account for 23–35% of the Rbfox2-expressing population between 4 and 7 dpi (Fig. 7E), with no significant difference observed among post-injury time-points. The Rbfox2-positive, myog:H2B-mRFP-negative cells we observe (see Fig. 7F′–H′ for magnified example at 4 dpi) likely include a combination of Pax7-positive satellite-like cells (Fig. 5F–G; Fig. S7A–C) and other cell types. Together, these data show that although there is overlap in Rbfox2 and Rbfox1l expression during post-injury repair, there are temporal expression differences, with Rbfox2 expressed earlier and predominantly in Pax7-positive satellite-like cells and Rbfox1l expressed later in newly-forming myofibers.
Figure 7. A subset of Rbfox2-expressing cells are myog:H2B-mRFP-positive during skeletal muscle repair.
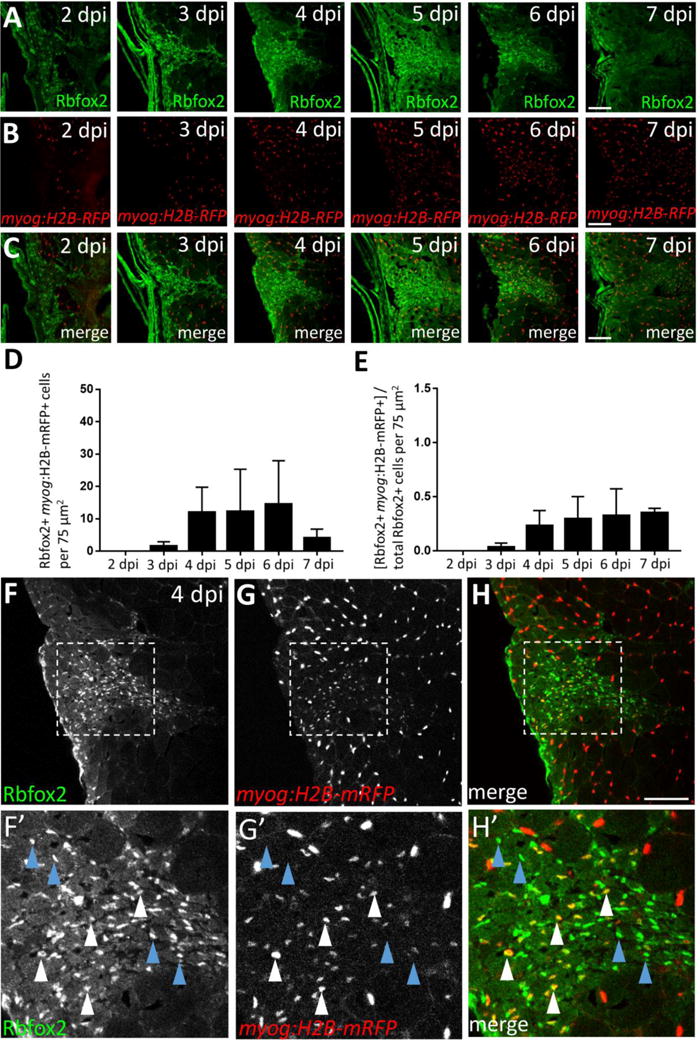
(A) Injury time-course experiment performed in myog:H2B-mRFP transgenic line showing Rbfox2 expression (on alternating sections with those described in Fig. 6). (B) As shown in Fig. 6B, myog:H2B-mRFP expression is readily observed within the injury site by 4 dpi. (C) Overlay of Rbfox2 and myog:H2B-mRFP expression. (D) Total number of Rbfox2, myog:H2B-mRFP double-positive cells at 2–7 dpi. (E) Ratio of Rbfox2-positive cells that are myog:H2B-mRFP-positive at 2–7 dpi. One-way ANOVA followed by Tukey’s multiple comparisons test was performed for all of the above statistical analyses; differences among time-points are not statistically significant. (F–H) Higher magnification view of 4 dpi injury site shown in A–C. (F′–H′) Area shown is the boxed region in F–H that highlights examples of Rbfox2-expressing cells that are myog:H2B-mRFP-positive (white arrowheads) and myog:H2B-mRFP-negative (blue arrowheads). Scale bar in all panels is 75 μm.
New myofibers that form after injury in adult zebrafish skeletal muscle may be derived from Pax7-positive satellite-like cells
Because murine satellite cells proliferate in response to injury and give rise to new muscle during skeletal muscle regeneration (Bentzinger et al., 2012; Montarras et al., 2013), we investigated whether adult zebrafish satellite-like cells give rise to new myofibers during injury-induced repair. We used the Tg(pax7a:GFP)i131 BAC transgenic line, which drives GFP expression under control of pax7a regulatory elements (Seger et al., 2011), in combination with Pax7 antibody labeling, to infer lineage relationships between satellite-like cells and newly-forming myofibers (Fig. 8). As expected, we find satellite-like cells at the injury site expressing both pax7a:GFP and Pax7 protein at 5 dpi (Fig. 8B–B″, arrowheads point to examples). Other cells express GFP but no Pax7 protein (Fig. 8B–B″); these pax7a:GFP-positive cells also express Myosin heavy chain (MyHC) (Fig. 8C–E; shown at 4 dpi), which is robustly expressed in newly-forming, small diameter myofibers. Detection of pax7a:GFP in newly-forming myofibers (in the absence of Pax7 protein) suggests that pax7a-driven GFP may perdure in satellite-like cells that give rise to new myofibers. We observe a similar pattern in the pax3a:GFP transgenic line at 4 dpi (Fig. 8F), with weaker GFP expression in newly-forming myofibers that overlaps with differentiation marker myog:H2B-mRFP (Fig. 8F; white arrowheads). Additionally, we note that most of the newly forming myofibers in adult zebrafish have peripheral nuclei, and only in rare cases is central nucleation observed (Fig. 8F″, blue arrowhead). These observations implicate satellite-like cells as a source of new muscle during adult zebrafish skeletal muscle repair.
Figure 8. GFP perdurance in the pax7a:GFP transgenic line implicates satellite-like cells as a source of new muscle fibers during injury-induced repair.
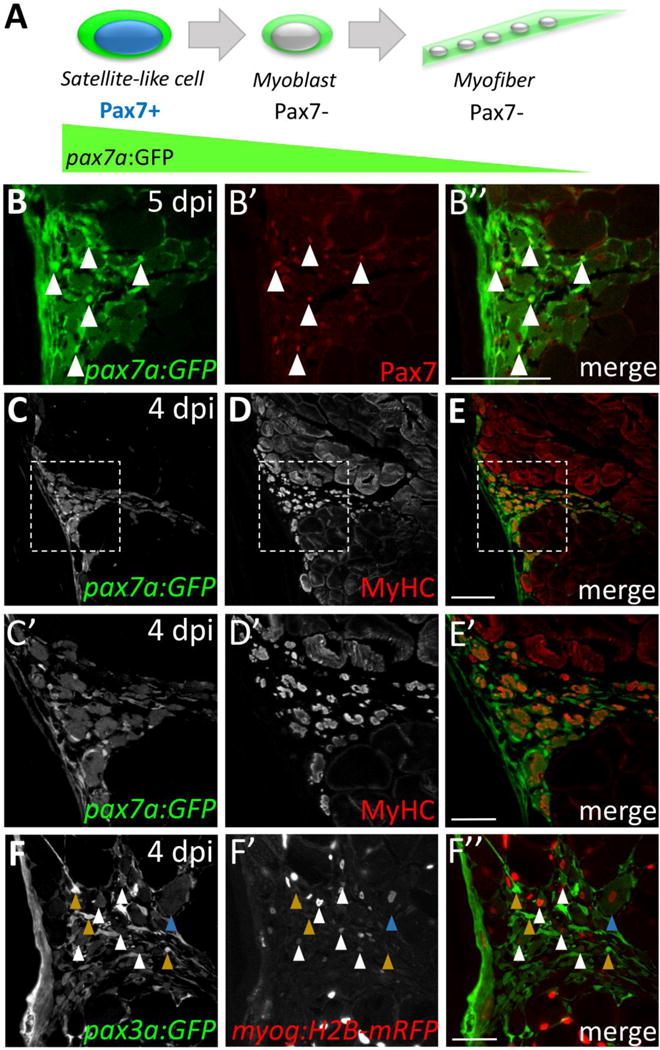
(A) Schematic diagram depicting Pax7 protein and pax7a:GFP transgene expression in satellite-like cells and myofibers. pax7a:GFP-positive; Pax7-positive cells represent satellite-like cells. Cells that show pax7a:GFP expression within the cytoplasm but are Pax7-negative are likely differentiating or differentiated cells in which pax7a-driven GFP perdures. (B–B″) At 5 dpi, pax7a:GFP and Pax7 double-positive satellite-like cells are present at the injury site (arrowheads); however, GFP expression is also detected in Pax7-negative presumptive newly-forming myofibers. (C–E) At 4 dpi, many cells expressing MyHC (A4.1025), a differentiation marker, also express pax7a:GFP. (C′–E′) Magnified view of boxed region in C–E highlights the overlap of pax7a:GFP and MyHC expression. (F–F″) pax3a:GFP and myog:H2B-mRFP transgene expression overlap in presumptive newly-forming myofibers at 4 dpi (white arrowheads), but pax3a:GFP-positive, myog:H2B-mRFP-negative are also present (mustard arrowheads). A rare presumptive newly-forming myofiber with a centrally-located nucleus is indicated by a blue arrowhead. Scale bar in B–E is 75 μm, in C′, D′, E′ and F–F” is 30 μm.
Pax7 is required for skeletal muscle repair in adult zebrafish
Given that Pax7 is necessary for proper muscle development and regeneration in mammals (Gunther et al., 2013; Oustanina et al., 2004; Seale et al., 2000; von Maltzahn et al., 2013), we investigated whether Pax7 is required for adult zebrafish muscle repair. Due to a teleost-specific genome duplication event, the zebrafish has two pax7 genes: pax7a and pax7b. Because pax7a and pax7b are both expressed in larval and/or adult zebrafish satellite-like cells and are implicated in repair and/or regeneration (Devoto et al., 2006, Feng et al., 2006, Gurevich et al., 2016; Hollway et al., 2007, Knappe et al., 2015; Li et al., 2013; Nord et al., 2013; Otten et al., 2012; Pipalia et al., 2016; Seger et al., 2011; Siegel et al., 2013; Stellabotte et al., 2007; Windner et al., 2015; this study), we made loss-of-function mutations in both genes (Fig. 9A, B). pax7a frame-shifting mutants (pax7aoz19 and pax7aoz23) are Pax7 protein null by immunohistochemistry (Fig. S8A–B), are viable, and show no obvious defects in skeletal muscle repair at 5 dpi (Fig. S8C–D). Similarly, pax7boz32 homozygous mutants display no significant difference in new myofiber formation at 4 dpi, compared to wild-type siblings (Fig. S9). To fully investigate Pax7 function in zebrafish muscle repair, we generated the pax7aoz23; pax7boz32 double mutant. pax7aoz23; pax7boz32 double mutants have pigmentation defects and survive to adulthood, consistent with recently published observations (Nord et al., 2016). Using the needle-stick injury paradigm, we find dramatic defects in adult skeletal muscle repair in pax7a; pax7b double mutants (Fig. 9). At 4 dpi, newly-forming MyHC-positive, Rbfox1l-positive myofibers are readily apparent at the injury site in wild-type adults (Fig. 9C; pax7a+/−; pax7b+/− sibling control). In contrast, we observe a near-complete absence of Rbfox1l and MyHC labeling at the injury site in pax7a−/−; pax7b−/− double mutants (Fig. 9D). We counted myofibers in 75 μm2 regions within the injury site and find on average only 4 newly-forming myofibers in pax7a−/−; pax7b−/− double mutants compared to an average of 45 newly-forming myofibers per 75 μm2 within the injury site in pax7a+/−; pax7b+/− sibling controls (n=2 adults per genotype; p<0.01, Student’s t-test; 176 total myofibers counted in pax7a+/−; pax7b+/−, 15 total myofibers counted in pax7a−/−; pax7b−/−). These data reveal that Pax7 function is required for adult zebrafish skeletal muscle repair.
Figure 9. Pax7 function is required for skeletal muscle repair in adult zebrafish.
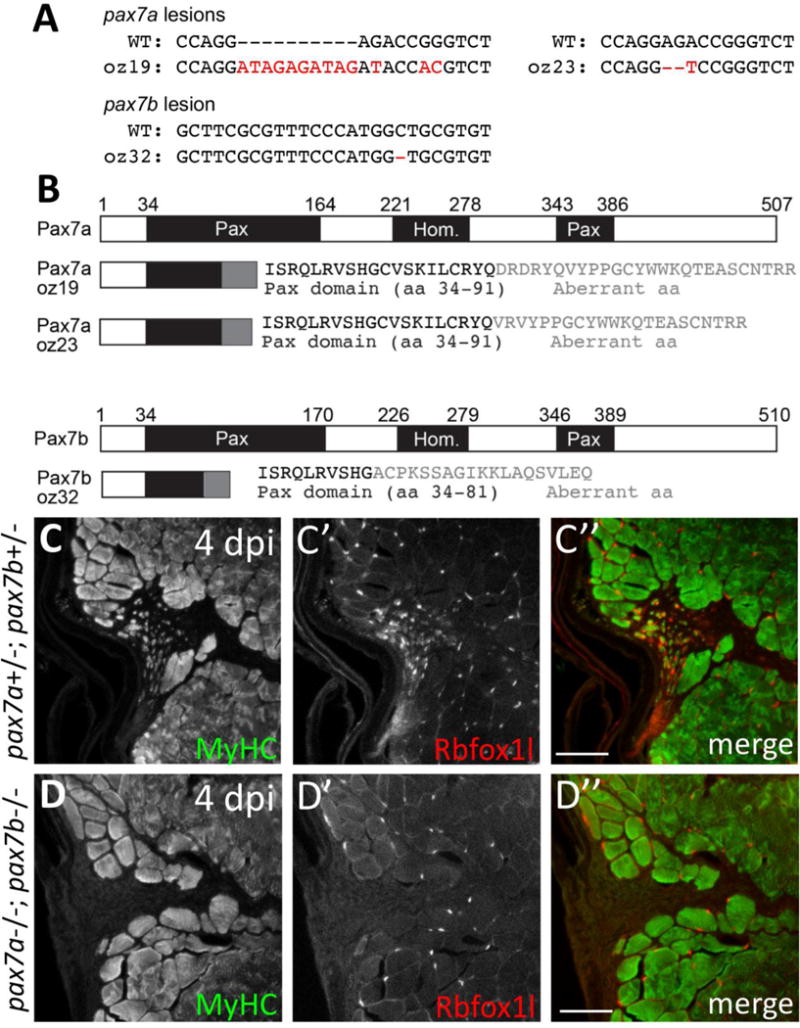
(A) DNA lesions for pax7a alleles (oz19, oz23) and the pax7b (oz32) allele. Mutant allele sequence is shown below the wild-type sequence. Red letters and dashes indicate the lesion as well as single nucleotide changes near the CRISPR-induced frameshifting mutation. (B) CRISPR-induced frameshifting mutations in exon 2 of pax7a and pax7b cause truncations in Pax7a and Pax7b proteins, respectively, that eliminate much of the first conserved paired domain (Pax) and the entire homeodomain (Hom) and second Pax domain. The pax7aoz19 and pax7aoz23 alleles encode the first 91 of 507 amino acids of the Pax7a protein, as well as an additional 26 and 22 aberrant amino acids, respectively. The pax7boz32 allele encodes the first 81 of 510 amino acids of the Pax7b protein, as well as an additional 19 aberrant amino acids. (C–C”) pax7a+/−; pax7b+/− doubly heterozygous sibling controls show normal muscle repair at 4 dpi, indicated by expression of newly-forming myofiber markers, MyHC (A4.1025) and Rbfox1l. (D–D”) In contrast, pax7a−/−; pax7b−/− double homozygous mutants are severely deficient in newly-forming myofibers at 4 dpi, indicated by the near absence of MyHC (A4.1025) and Rbfox1l expression within the injury site. The pax7aoz23 and pax7boz32 alleles were used in these experiments. Scale bar in all panels is 75 μm.
DISCUSSION
Muscle injury induces satellite-like cell activation and muscle repair in adult zebrafish skeletal muscle
The discovery of Pax7 as a reliable marker for satellite cells in mouse (Seale et al., 2000) has allowed investigators to examine whether similar cells are present in other model systems. Building on the identification of Pax7-positive satellite-like cells in larval and adult zebrafish (Berger et al., 2010; Hollway et al., 2007; Knappe et al., 2015; Seger et al., 2011; Siegel et al., 2013; Tee et al., 2012; Zhang and Anderson, 2014), we investigated whether adult zebrafish possess satellite-like cells that function in skeletal muscle repair. We find cells situated within the basal lamina that contain dense heterochromatin, express Pax7, proliferate in response to injury, and likely give rise to new myofibers during adult skeletal muscle repair, all properties of satellite cells in mammals. Additionally, we have identified that Rbfox2 and Rbfox1l are expressed predominantly in satellite-like cells and newly-forming myofibers, respectively, during the muscle repair process (Fig. 10A, B). Overall, we find Pax7, pax3a:GFP and Rbfox2 are expressed and EdU is incorporated during the time of progenitor cell expansion and proliferation, while Rbfox1l and myog:H2B-mRFP are expressed during later stages of myofiber differentiation during adult zebrafish skeletal muscle repair (Fig. 10B). A summary of markers and their relative expression (or incorporation, for EdU) levels during the post-injury time-course is depicted in Fig. 10B. Our findings illustrate that the zebrafish is a suitable vertebrate model system to study adult satellite cells and skeletal muscle repair.
Figure 10. Model depicting changes in gene expression as satellite-like cells differentiate into new muscle fibers during adult zebrafish skeletal muscle repair.
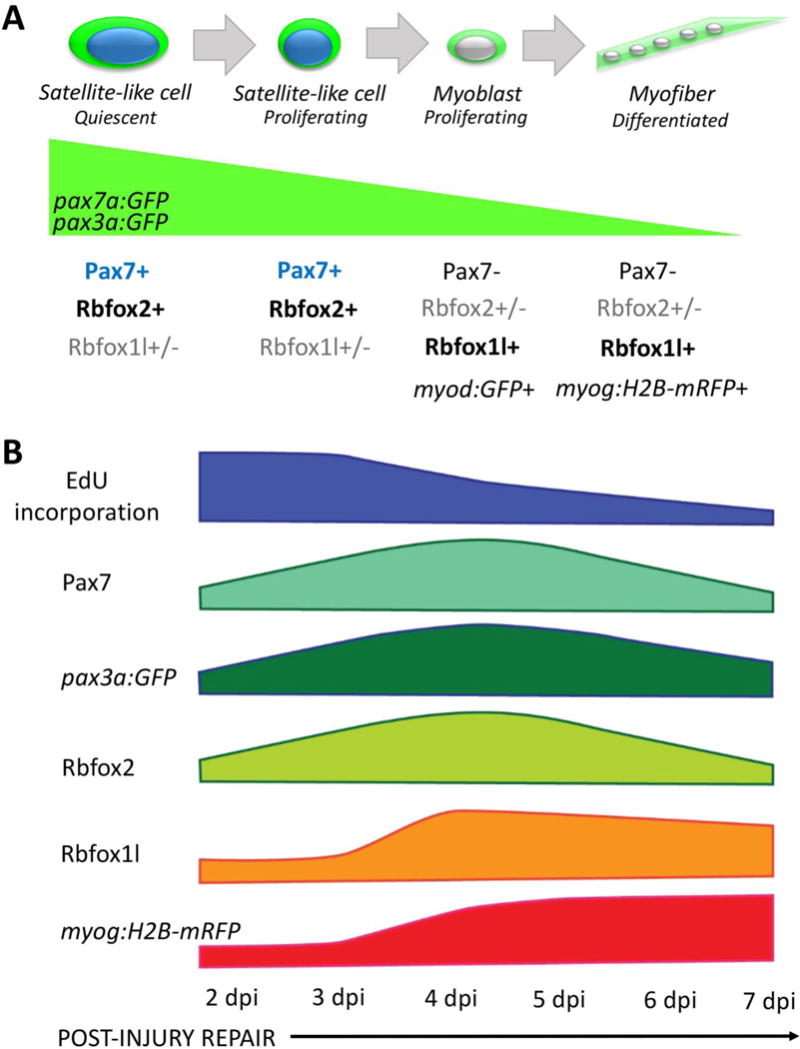
(A) Adult zebrafish satellite-like cells express Pax7 and Rbfox2, and in transgenic pax7a:GFP and pax3a:GFP adults, express GFP. A subset (~37%) express Rbfox1l. During injury-induced repair, some Pax7-expressing satellite cells-like become activated; initially, activated cells proliferate, then down-regulate Pax7 and activate differentiation markers. Although Pax7 protein is largely absent in myoblasts, pax7a-driven and pax3a-driven GFP perdures in pax7a:GFP and pax3a:GFP transgenic lines (green wedge). As cells adopt more differentiated fates, Rbfox1l becomes prominently expressed, as do transgenic markers of myoblasts (myod:GFP) and myofibers (myog:H2B-mRFP). (B) Diagram depicting marker expression and proliferation status at the population level at the injury site at 2–7 dpi. A large number of proliferating (EdU+) cells are present at the injury site at 2 and 3 dpi, some of which are Pax7-positive. This early proliferation of Pax7-positive satellite-like cells appears to lead to an increase in Pax7-positive cell numbers by 4 dpi. pax3a:GFP shows a similar wave of expression that peaks at the time when Pax7-positive cell numbers are highest; however, pax3a:GFP expression is also observed at later post-injury time-points (as is pax7a:GFP expression). Rbfox2 expression largely mirrors that of Pax7, although Rbfox2 is expressed in some myonuclei of new myofibers that no longer express Pax7. Rbfox1l is robustly expressed at the onset of new myofiber formation (3–4 dpi), with peak expression at 4 dpi in both nucleus and cytoplasm of newly-forming fibers. The number of myog:H2B-mRFP-positive cells gradually increase post-injury as myogenic differentiation and new myofiber formation progresses.
Zebrafish adult muscle repair shares similarities with larval skeletal muscle repair
Previous studies in larval stage zebrafish revealed a population of Pax7-positive satellite-like cells (Berger et al., 2010; Knappe et al., 2015; Seger et al., 2011). Additionally, pax7a-driven GFP expression has been detected in cells concentrated at injury sites within 24–48 hours post injury of larval zebrafish at 4–5 days post-fertilization (dpf) (Knappe et al., 2015; Seger et al., 2011) and 7 dpf (Knappe et al., 2015). Similarly, we show that Pax7-expressing cells in adult zebrafish muscle increase after injury, peaking at 4 dpi and then declining sharply; this brief window of satellite cell expansion was not described in a previous adult study (Tee et al., 2012) because the time-point examined (7 dpi) occurs after the transient period of satellite cell expansion. Also similar to our adult findings, larval myofibers express pax7a:GFP after cardiotoxin injury at 4–5 dpf (Seger et al., 2011) and after extensive needle-stick injury (using a wider-bore needle) at 4 and 7 dpf (Knappe et al., 2015), suggesting that pax7a-expressing zebrafish satellite-like cells contribute to new myofiber formation after injury at both larval and adult stages. A previous study found that a large injury (in contrast to small focal injury) is needed to induce pax7a:GFP expression in myofibers during larval muscle repair (Knappe et al., 2015). Although it is possible that our adult needle stick paradigm causes more extensive injury than focal injury in larval fish (Knappe et al., 2015), we note that the pax7a:GFP transgenic line used by Knappe and colleagues (2015) is distinct from the line used in the cardiotoxin study (Seger et al., 2011) and our current study. Future adult injury studies in both transgenic lines, that incorporate both small and large injuries will aid in addressing potential differences. In all these studies, GFP perdurance in newly-forming myofibers was used to infer that they are derived from pax7a:GFP-expressing satellite-like cells, thus differences between lines, injury paradigms, or adult and larval programs, could contribute to differences among studies. CreER-based lineage tracing approaches will more definitively address the extent to which Pax7-expressing cells contribute to new muscle after injury.
Injury-induced skeletal muscle repair in adult zebrafish: the use of stem/progenitor cells versus dedifferentiation
Our findings indicate that adult zebrafish possess satellite-like cells that function like mammalian satellite cells during skeletal muscle repair. Although pax7-driven GFP protein perdurance supports the idea that satellite-like cells contribute to new muscle after injury, it is not a definitive demonstration, and it is possible that new muscle forms by satellite cell-independent mechanisms. Some adult zebrafish tissues, like heart, fin, extraocular muscle, and retina, regenerate via dedifferentiation of mature cell types without using a specific precursor population (Geurtzen et al., 2014; Jopling et al., 2010; Knopf et al., 2011; Poss et al., 2002; Ramachandran et al., 2010; Saera-Vila et al., 2015; Saera-Vila et al., 2016; Sousa et al., 2011; Stewart et al., 2012; Wan et al., 2012). Particularly relevant to our study, post-injury regeneration of adult extraocular skeletal muscle uses dedifferentiation (Saera-Vila et al., 2015; Saera-Vila et al., 2016), demonstrating that some skeletal muscle types are repaired using satellite cell-independent mechanisms. However, another study using Cre/lox-based lineage tracing approaches found that dedifferentiation does not occur during regeneration of larval zebrafish tail muscle following amputation (Rodrigues et al., 2012). Additionally, recent studies have described larval zebrafish trunk muscle progenitor cell populations that contribute to new muscle fibers during repair after needle-stick injury (Gurevich et al., 2016; Pipalia et al., 2016). Using lineage tracing, Gurevich and colleagues (2016) found no evidence for dedifferentiation during larval trunk skeletal muscle repair. Our work strongly suggests that a local satellite-like cell population contributes to adult zebrafish skeletal muscle repair, but future lineage tracing studies will provide definitive evidence.
Pax3-expressing cells also contribute to muscle repair in both fish and mice
In this study, we have shown that zebrafish pax3a:GFP-positive satellite-like cells, like pax7a:GFP-positive satellite-like cells, are induced at the site of muscle injury and likely contribute to the repairing myotome. Because baseline pax3a:GFP transgene expression in larvae and adults is different, it is difficult to directly compare our results to a study performed in pax3a:GFP transgenic larval fish (Seger at el., 2011). In our study, the number of pax3a:GFP-expressing cells is very low in uninjured adult muscle and robustly increases after injury; by contrast, there are already many pax3a:GFP-expressing cells in uninjured larval skeletal muscle and further increases are not observed after cardiotoxin-induced muscle injury (Seger et al., 2011). Additional larval studies are needed to distinguish whether the difference reflects variation in regenerative potential of pax3a:GFP-expressing cells or in differential expansion of the population.
In mammals, Pax3 is differentially expressed in satellite cells associated with different muscles; the mouse diaphragm contains more Pax3-expressing cells than Pax7-expressing cells, whereas tibialis anterior (TA) satellite cells primarily express Pax7 and not Pax3 (Kassar-Duchossoy et al., 2005; Montarras et al., 2005; Relaix et al., 2005). Coupled with knockout studies (Gunther et al., 2013; Relaix et al., 2005; Seale et al., 2000; von Maltzahn et al., 2013), the idea emerged that Pax3 and Pax7 may have both overlapping and distinct functions in murine satellite cells. In adult zebrafish, we find Pax7-expressing and pax3a:GFP-expressing cells in injured muscle, some of which express both Pax7 and pax3a:GFP (94% of Pax7-positive cells are also pax3a:GFP-positive), whereas others express only Pax7 or pax3a:GFP. Using GFP perdurance in pax7a:GFP and pax3a:GFP transgenic lines, we infer that both pax7a:GFP- and pax3a:GFP-expressing cells can generate new myofibers after injury; however, whether there is an independent population of Pax3-positive (but Pax7-negative) satellite-like cells that contributes to muscle repair after injury remains to be determined. As little is currently known about satellite cell sub-populations and whether they are differentially involved in skeletal muscle repair, future studies may provide novel insight into satellite cell heterogeneity and function.
Satellite-like cell localization in the zebrafish myotome and implications for the muscle stem cell niche and fiber type repair and regeneration
We have shown that adult zebrafish satellite-like cells are enriched within uninjured slow muscle, at the horizontal myoseptum, and at the boundary between fast and slow muscle domains. The differential localization of satellite-like cells is readily apparent due to the well-described spatial segregation of slow and fast muscle domains of the zebrafish myotome (Bassett and Currie, 2003; Devoto et al., 1996; Elworthy et al., 2008; Jackson et al., 2015; Ju et al., 2003; Nord et al., 2014; Waterman, 1969). In the zebrafish larval myotome, pax7a-expressing satellite-like cells are also concentrated largely around the edges of the slow muscle domain and at the horizontal myoseptum (Hollway et al., 2007; Knappe et al., 2015; Seger et al., 2011). We hypothesize that these areas represent a satellite-like cell niche. Intriguingly, previous work in the mouse also reported that more satellite cells associate with slow versus fast fibers (Collins et al., 2005; Shefer et al., 2006). Does the preferential association of satellite cells with slow muscle fibers have implications for regenerative capacity of slow versus fast fibers? Do resident satellite cells give rise to slow and fast muscle equivalently, or is there a hierarchical process (Wang et al., 2015)? Finally, do pax7a and/or pax3a-expressing satellite-like cell populations differentially contribute to fast and slow muscle? Studies that employ lineage-tracing methods and fiber type-specific transgenic lines will further investigate the relationship between satellite cell sub-populations and muscle fiber type regeneration.
Functional implications for Rbfox1l and Rbfox2 in skeletal muscle repair
It has long been appreciated that many molecular mechanisms regulating development of embryonic cells into differentiated tissue types are re-utilized during tissue regeneration and repair in adults. In this work, we show that Rbfox2 and Rbfox1l protein expression in the zebrafish adult myotome parallels what we previously described for rbfox2 and rbfox1l transcript expression in embryos (Gallagher et al., 2011), with Rbfox2/rbfox2 expression higher in less differentiated myogenic progenitor cells and Rbfox1l/rbfox1l expression increasing as cells differentiate into muscle. We hypothesize that Rbfox proteins regulate a muscle-specific splicing program that is employed first during embryonic muscle development and later in adult skeletal muscle satellite cells. Based on their expression patterns, we propose that Rbfox2 promotes satellite-like cell specification and/or maintenance, while Rbfox1l promotes satellite-like cell differentiation.
Do Rbfox proteins function in muscle regeneration and repair? RBFOX2 is required for human embryonic stem cell survival (Yeo et al., 2009) and promotes differentiation of human induced pluripotent stem cells (Venables et al., 2013). While Rbfox2 is expressed in many mammalian cell types including muscle, blood, embryonic, and neuronal cells (Dietrich et al., 2015; Underwood et al., 2005; Yeo et al., 2007), most Rbfox2 studies have focused on neuronal function (Gehman et al., 2012; O’Brien et al., 2012; Underwood et al., 2005; Weyn-Vanhentenryck et al., 2015; Yeo et al., 2007), though one recent study focused on Rbfox2 function in the mouse heart (Wei et al., 2015). However, whether Rbfox2 is expressed in mouse satellite cells, and/or functions during skeletal muscle development, growth, and/or regeneration has not been studied. The closest murine Rbfox1l homolog, Rbfox1, is primarily studied as a regulator of neuronal splicing (Gehman et al., 2011; Hamada et al., 2016; O’Brien et al., 2012; Weyn-Vanhentenryck et al., 2015), but is also expressed in heart and skeletal muscle (Kalsotra et al., 2008; Kiehl et al., 2001; Underwood et al., 2005). Surprisingly, conditional and partial Rbfox1 depletion in satellite cells does not affect muscle regeneration, with the caveat that only ~70% knockdown was achieved (Pedrotti et al., 2015). Recent work in C2C12 cells has identified a requirement for Rbfox1 and Rbfox2 in myotube differentiation in vitro (Runfola et al., 2015). Future studies in satellite cells in vivo and during regeneration may shed light on potential roles for Rbfox RNA-binding proteins in satellite cell regulation and skeletal muscle regeneration in vertebrates.
Do Rbfox proteins have non-nuclear functions?
We observe strong cytoplasmic Rbfox1l expression in newly-forming myofibers, which is intriguing since Rbfox proteins have been largely studied as regulators of alternative splicing, a nuclear function. We also observe cytoplasmic Rbfox2 in what appear to be non-muscle cells located between uninjured muscle fibers. Similarly, cytoplasmic Rbfox expression has been observed in neural tissue (Hamada et al., 2013; Lee et al., 2016), in germ cells (Carreira-Rosario et al., 2016), and various cell lines (Lee et al., 2009; Nakahata and Kawamoto, 2005), and a recent study has identified a role for cytoplasmic Rbfox1 in cortical development (Hamada et al., 2015). Since rbfox transcripts themselves are also targets of alternative splicing (Baraniak et al., 2006; Damianov and Black, 2010; Dredge and Jensen, 2011; Lee et al., 2009; Nakahata and Kawamoto, 2005; Yang et al., 2008), it is possible that cytoplasmic Rbfox1l reflects post-transcriptional auto-regulation that tunes the level of Rbfox1l-regulated splicing events in the nucleus. Alternatively, or perhaps additionally, Rbfox1l may have cytoplasmic function(s) that influence transcript stability, localization, and/or translation as has been described for other RNA binding proteins (RBPs) (Blech-Hermoni and Ladd, 2013; Darnell and Richter, 2012). There is precedent for RNA-binding proteins having dual roles in both the nucleus and cytoplasm, as has been reported for Drosophila ELAV proteins (HuB, C, and D) (Bolognani and Perrone-Bizzozero, 2008; Fukao et al., 2009; Zhu et al., 2006). Indeed, recent work shows that RBFOX1/Rbfox1 may promote mRNA stability in mammals (Ray et al., 2013; Lee et al., 2016); for example, cytoplasmic Rbfox1 has been shown to regulate expression of autism-associated genes by binding neuronal transcript 3′ UTRs at sites that overlap significantly with miRNA binding sites (Lee et al., 2016). Additionally, regulation by Rbfox1 has been observed at the level of translation, whereby Drosophila Rbfox1 represses translation of transcripts containing 3′UTR Rbfox binding elements (Carreira-Rosario et al., 2016). Interestingly, Rbfox2 can regulate miRNA biogenesis by binding to miRNA precursors in mammalian cells (Chen et al., 2016). Taken together, we conclude that Rbfox proteins have important and conserved cellular regulatory functions in both the nucleus and cytoplasm.
Is Pax7 function in adult skeletal muscle repair conserved among vertebrates?
Several studies have established that Pax7 is required for proper adult skeletal muscle regeneration (Gunther et al., 2013; Oustanina et al., 2004; von Maltzahn et al., 2013), though another study indicated that Pax7 is dispensable (Lepper et al., 2009). The former studies found that regeneration was impaired after cardiotoxin injury in Pax7 mutants, especially after multiple rounds of injury (Gunther et al., 2013; Oustanina et al., 2004; von Maltzahn et al., 2013). In our study, we have shown that pax7a; pax7b double mutant zebrafish are defective in adult skeletal muscle repair, revealing that pax7 function is conserved during skeletal muscle regeneration in mice and zebrafish.
Is impaired regeneration in Pax7 knockout mice due to a diminished satellite cell population or a diminished ability of satellite cells to participate? Pax7 is required for satellite cell maintenance in adult mice, as conditional knockout of Pax7 results in decreasing numbers of satellite cells over time (Gunther et al., 2013; von Maltzahn et al., 2013). However, it is unclear from these studies whether the few remaining satellite cells are able to proliferate and mount a satellite cell response post-injury, as satellite cell numbers were assessed at later post-injury time-points when regeneration is normally complete. Intriguingly, a previous study has reported that Pax7-lacZ-positive cells actually increase after injury in adult Pax7 knockout mice (Oustanina et al., 2004), suggesting that satellite cells may be able to proliferate post-injury, but are unable to properly differentiate. Whether Pax7 is required for proliferation and/or maintenance of satellite cells or for satellite cell differentiation remains to be confirmed. Future work in both mice and fish will resolve which aspects of regeneration are controlled by Pax7.
EXPERIMENTAL PROCEDURES
Zebrafish Strains and Maintenance
Zebrafish strains were maintained according to The Ohio State University Institutional Animal Care and Use Committee (IACUC). Adult zebrafish between 2 and 6 months of age were used in our experiments, except in Figure 4E–E″ where the fish were 12 months old. Three to five adult zebrafish (of random sex) were used in experiments shown in Figs. 1, 2, and 8. Two adult zebrafish (of random sex) were used for each time-point and/or genotype in injury time-course experiments (Figs. 3, 5, 6, 7 and supplements to these figures), pax7a; pax7b double mutant analysis (Fig. 9, S8, S9), and Fig. 4 A–D (3 adults were analyzed in Fig. 4E). Zebrafish lines used were the AB wildtype strain, the BAC transgenic lines TgBAC(pax3a:GFP)i150 and TgBAC(pax7a:GFP)i131 (Seger et al., 2011), the dmdct90aGt transgenic insertion line that expresses Dystrophin-Citrine fusion protein (Trinh et al., 2011) and is referred to as Dystrophin FlipTrap in our work, Tg(smyhc1:EGFP)i104 (Elworthy et al., 2008), Tg(mylpfa:mCherry)cz3327 (Ignatius et al., 2012), TgBAC(myod:GFP)i124 (Seger et al., 2011), Tg(-80.0myf5:EGFP)zf37 (Chen et al., 2007), Tg(acta1:GFP)zf13 (also known as alpha-actin:GFP; Higashijima et al., 1997), and Tg(myog:H2B-mRFP) (Tang et al., 2016). pax7aoz19, pax7aoz23, and pax7boz32 mutant zebrafish were generated in this study (see below). All mutant experiments were performed on zebrafish derived from F2 or later filial generation incrosses. The allele combination used for double mutant analysis was pax7aoz23; pax7boz32.
Needle-stick injury procedure
Needle-stick injury was performed as previously described (Rowlerson et al., 1997). Adult zebrafish were anesthetized with Tricaine (0.004% diluted in fish system water), and muscle injury was performing by inserting and withdrawing a 27 gauge needle (0.4 mm) into the ventral myotome of the post-anal tail. Ventral myotome injury was performed to avoid damaging satellite-like cell populations at the horizontal myoseptum and the spinal cord. Zebrafish were returned to fresh water and monitored after injury. Zebrafish lacking full recovery (e.g. showing excessive bleeding and/or lethargic swimming) were euthanized. Injured zebrafish were housed and fed per normal protocols for 2 to 7 dpi until analysis.
Rbfox1l and Rbfox2 antibody production
Using Genomic Antibody Technology (SDIX LLC), polyclonal antibodies were raised in rabbits against divergent N-terminal regions of zebrafish Rbfox1l and Rbfox2. For anti-Rbfox1l, an immunogen of 81 amino acids spanning positions 30–110 was affinity purified and validated by ELISA against recombinant Rbfox1l protein. For anti-Rbfox2, an immunogen of 80 amino acids spanning positions 34–113 was affinity purified and validated by ELISA against recombinant Rbfox2 protein. Immunogenic peptides are:
Rbfox1l Immunogen sequence: GAAAQEAGPGNGDPSLPQVYAPPPSYPPPGQAPPTPAARLPPLDFSAAHPNSEYADHHQLRV YQGPQHDGTESITASNTDD
Rbfox2 Immunogen sequence: PPPQNGLSLEFSSGGGGSSVYSSSGQTEAVAGTSSGASNPSTQLSDASPQPDNQLVLVSNAV AIREDSSEAKGTPKRLHV
Rbfox1l and Rbfox2 antibody validation
Zebrafish Rbfox1l and Rbfox2 polyclonal antibodies were validated by immunoblot analysis of embryo lysates from uninjected wild-type and Rbfox1l- and/or Rbfox2-depleted morphant embryos. To obtain Rbfox-depleted morphant samples, rbfox1l and rbfox2 splice-blocking morpholinos, 5′-GCATTTGTTTTACCCCAAACATCTG-3′ and 5′-TATAATGCTTTATATACCCCGAACA-3′, respectively, were injected into the yolk of 1-cell stage embryos as previously described (Gallagher et al., 2011) and raised at 28.5°C for 24 hours. Zebrafish embryos were manually dechorionated at the 24 hpf stage and rapidly pipetted in cold Cell Resuspension Buffer (116 mM NaCl, 2.9 mM KCl, 5.0 mM HEPES, pH 7.2, 0.1 mg/mL Soybean Trypsin Inhibitor, Complete Protease Inhibitor Cocktail [Roche]). Cell lysis was performed in 2X LDS Buffer (Life Technologies). Cell extract equivalent to 2 embryos was loaded per lane, electrophoresed, and transferred to PVDF membrane using the Iblot semi-dry transfer system (Invitrogen). The blot was first probed with anti-Rbfox2 primary antibody (1:2500) and AP-conjugated goat anti-rabbit secondary antibody (1:10,000), developed, stripped according to the Western Superstar protocol, and then re-probed with anti-Rbfox1l primary antibody (1:2500) and AP-conjugated goat anti-rabbit secondary antibody (1:10,000). Blots were developed using the Western Superstar Kit (Applied Biosystems) according to manufacturer protocol. A separate blot loaded with the same protein lysates was processed at the same time, probed with anti-alphaTubulin primary antibody (1:2500) and AP-conjugated anti-mouse secondary antibody (1:10,000).
pax7a and pax7b mutant generation
pax7a and pax7b zebrafish mutants were generated using previously described CRISPR methods (Talbot and Amacher, 2014). For pax7a, a guide RNA (gRNA) targeting Exon 2 was synthesized from the following CRISPR target sequence: GGCGGATAGACCCGGTCTCC. gRNA and Cas9 mRNA were co-injected into one-cell stage zebrafish embryos. Injected embryos were grown to adulthood, and progeny from wild-type outcrosses were screened to identify germline-transmitting founders. Two frame-shifting alleles were generated: oz19 (10 base-pair insertion) and oz23 (2 base-pair deletion) (see Fig. 9A). Genotyping for oz19 was performed using pax7a forward 5′-TATCCGACCCTGTGTTATCTCCAGAC-3′ and reverse 5′-AATCGTTCACTCACCCTCGGTTTG-3′ primers. The pax7aoz19 amplicon (147 bp) is distinguished from the wild-type amplicon (137 bp) by size on an agarose gel. Genotyping for oz23 was performed using pax7a forward 5′-GGTCAAGGTCGAGTCAATCAAC-3′ and reverse 5′-AGCTGAACATCCCTGGATTCTC-3′ primers. AvaII digestion cuts the pax7aoz23 amplicon into two products (300 bp, 176 bp), whereas the wild-type amplicon is not cut (478 bp). For pax7b, a guide RNA targeting Exon 2 was synthesized from the following CRISPR target sequence: GGATTTTGGACACGCAGCCA. A 1 bp frame-shifting allele, oz32, was generated (see Fig. 9A). Genotyping for oz32 was performed using pax7b forward 5′-CATCAATGGCAGGCCTTTGC-3′ and reverse 5′-CACACCCCGTCCTTCAACAG-3′ primers. Fnu4HI digestion cuts the wild-type amplicon into two products (425 bp, 110 bp), whereas the pax7boz32 amplicon is not cut (534 bp).
Tissue processing and immunohistochemistry
Adult zebrafish were anesthetized with Tricaine (0.004% diluted in fish system water) and fixed overnight in 4% PFA by immersion fixation. The following day, tail skeletal muscle was isolated using a razor blade and washed in a glass vial containing 1X PBS for 10 minutes. Subsequently, 1X PBS solution was replaced with a solution of 30% sucrose in 1X PBS (cryopreservation), and samples were placed on a rocker for approximately 3 hours at room temperature until tissue samples sank to the bottom of vial. Muscle tissue was embedded in OCT (Sakura Finetek), frozen on a slab of dry ice, and cut using a Microm 550 cryostat into 25 μm thick cryosections onto Superfrost Plus Microscope slides (Thermo Fisher Scientific) and stored at −20°C. Sections of the injury area anterior to and including the needle lesion were collected. Immunohistochemistry was performed as previously described for adult zebrafish tissue sections (Berberoglu et al., 2009; Berberoglu et al., 2014). Briefly, slides were washed in 1X PBS at room temperature to rehydrate tissue and remove OCT, and then incubated in PBS + 0.5% Triton X-100 for 10 minutes to permeabilize membranes. Slides were washed in 1X PBS for 15 minutes, blocked in PBS + 3% bovine serum albumin (BSA) for 1–2 hours at room temperature, and incubated in primary antibody overnight at 4°C. The following day, slides were washed 2–3 times with 1X PBS over 1 hour, followed by PBS + 0.1% Triton X-100 for 10 minutes. Slides were then incubated in secondary antibody (diluted in PBS + 0.1% Triton X-100) for 2 hours at room temperature. Slides were washed 2–3 times for 30 minutes in 1X PBS, mounted with fluorescent mounting medium (Dako), and coverslipped. In our experiments, anti-GFP antibody was used to amplify GFP signal.
The following primary antibodies were used at the indicated dilutions: mouse anti-Pax7 (DSHB; 1:20); chicken anti-GFP (ab13970, Abcam; 1:2000), rabbit anti-Rbfox2 (1:1000), rabbit anti-Rbfox1l (1:1000), rabbit anti-Laminin (L9393, Sigma; 1:100), mouse anti-MyHC (A4.1025, DSHB; 1:1000). The following Alexa Fluor secondary antibodies were used at 1:200 dilution: anti-chicken 488, anti-rabbit and anti-mouse 488, 568, and 647 (Thermo Fisher Scientific).
Intraperitoneal EdU injection to label S-phase cells
For EdU labeling experiments, adult zebrafish were injected intraperitoneally following Tricaine anesthetization with 20 uL of 10 mM EdU (5-ethynyl-2′-deoxyuridine; Life Technologies) diluted in 1X PBS. Zebrafish were returned to fresh water after injection and monitored for recovery from anesthesia. Fish that did not regain consciousness were euthanized per IACUC protocol. For EdU pulse experiments (acute EdU labeling), zebrafish were housed for 4 hours to allow for EdU incorporation before fixation and tissue processing as described above. For EdU labeling on tissue sections, the click-chemistry reaction was performed as described (Invitrogen Life Technologies). Following PBS + 0.5% Triton incubation and 15 minute 1X PBS wash (described above), a 10 minute incubation of EdU labeling solution (in the dark) is the optimal labeling time used in our experiments. Incubation is followed by a 15-minute wash with 1X PBS before blocking.
Transmission Electron Microscopy
Transmission electron microscopy was performed according to standard procedures set by the OSU Campus Microscopy & Imaging Facility. Methods were adapted for adult zebrafish skeletal muscle as follows. In brief, adult zebrafish were anesthetized and then decapitated to sacrifice. Skin was removed from tail muscle region of interest, and a transfer pipette was used to place a few drops of fixative (2% glutaraldehyde in 0.1M phosphate buffer, pH 7.4) onto the tissue. After a few minutes, tail muscle tissue was dissected out and fixed (same fixative) an additional 15 minutes (in a small dish). Tissue from both slow and fast-twitch muscle fiber domains was cut into small pieces (1mm × 2mm, to aid in orientation, using a fine scalpel) and fixed in vials overnight at 4°C. Samples were embedded using standard procedures for TEM, and thick sections were taken and stained with toluidine blue. Orientation was assessed, and thin sections (70–90 nm; cross-sections) were then taken from a smaller area of the block for labeling and imaging.
Confocal imaging, image processing, and statistical analyses
Samples were imaged using a Leica TCS SL point-scanning laser confocal inverted microscope or an Andor Revolution WD spinning disk laser confocal microscope system (Inverted Nikon TiE microscope). Confocal projection images (~15μm or narrower) and single confocal z-sections (0.5–1 μm) were used to assess co-localization of markers. For quantification, all cells counted had at least one marker localized to the nucleus to ensure accurate cell counts. Cells were counted manually on 2 sections from each of 2–3 adult zebrafish per genotype and/or time-point using NIH ImageJ. Cell counts over 2 sections were averaged per adult zebrafish; percentages of cells are reported based on an average of averages (for statistical comparisons). Numbers of cells counted in experiments are reported as absolute numbers (without averaging between sections). Graphpad Prism 7 was used for statistical analysis. Adobe Photoshop CS6 and NIH ImageJ were used for image processing. Student’s t-test was performed for statistical comparison of two conditions. For injury time-course experiments, a One-Way ANOVA followed by Tukey’s multiple comparisons test was performed. p*<0.05, p**<0.01, p***<0.001.
Supplementary Material
HIGHLIGHTS.
-
✥
Cells similar to mammalian satellite cells are present in adult zebrafish skeletal muscle.
-
✥
Adult Pax7-positive satellite-like cells proliferate and contribute to skeletal muscle repair after injury.
-
✥
RNA-binding proteins Rbfox2 and Rbfox1l are expressed in satellite cells and regenerating myofibers, respectively, during skeletal muscle repair.
-
✥
Pax7 is required for skeletal muscle repair in adult zebrafish.
Acknowledgments
This work was supported by intramural funds from the Ohio State University and Nationwide Children’s Hospital Center for Muscle Health and Neuromuscular Disorders (CMHND), NIH grant R01GM117964 (to S.L.A.), and American Heart Association Grant #0825194F (T.L.G.). M.A.B. was supported by a CMHND-administered NIH/NINDS T32 training grant (NS077984) and by a Pelotonia Postdoctoral Fellowship. We thank Phillip Ingham for pax7a:GFP, pax3a:GFP, and myod:GFP BAC transgenic lines and for the smyhc1:EGFP line, and Le Trinh and Scott Fraser for the Dystrophin FlipTrap line. We thank Richard Montione and the OSU Campus Microscopy & Imaging Facility for TEM assistance, the Neuroscience Imaging Core for equipment and advice, Rightmire fish facility staff for excellent fish maintenance, Brittany Siegenthaler and Anne Witzky for genotyping assistance, and Anne Witzky and Duy Phan for antibody testing. Finally, we thank Amacher lab members, the CMHND “Muscle Group” research community, and Rightmire Zebrafish Groups for helpful discussions throughout the course of this study, and Denis Guttridge for reading and commenting on an early draft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asakura A, Komaki M, Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–253. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DI, Currie PD. The zebrafish as a model for muscular dystrophy and congenital myopathy. Hum Mol Genet. 2003;12:R265–270. doi: 10.1093/hmg/ddg279. [DOI] [PubMed] [Google Scholar]
- Baxendale S, Chen CK, Tang H, Davison C, Hateren LV, Croning MD, Humphray SJ, Hubbard SJ, Ingham PW. Expression screening and annotation of a zebrafish myoblast cDNA library. Gene Expr Patterns. 2009;9:73–82. doi: 10.1016/j.gep.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4(2) doi: 10.1101/cshperspect.a008342. doi:pii: a008342. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberoglu MA, Dong Z, Li G, Zheng J, Martinez LC, Peng J, Wagle M, Reichholf B, Petritsch C, Li H, Pleasure S, Guo S. Heterogeneously expressed fezf2 patterns gradient Notch activity in balancing the quiescence, proliferation, and differentiation of adult neural stem cells. J Neurosci. 2014;34:13911–13923. doi: 10.1523/JNEUROSCI.1976-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberoglu MA, Dong Z, Mueller T, Guo S. fezf2 expression delineates cells with proliferative potential and expressing markers of neural stem cells in the adult zebrafish brain. Gene Expr Patterns. 2009;9:411–422. doi: 10.1016/j.gep.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Berger S, Hall TE, Lieschke GJ, Currie PD. Dystrophin-deficient zebrafish feature aspects of Duchenne muscular dystrophy pathology. Neuromuscular Disorders. 2010;20:826–832. doi: 10.1016/j.nmd.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Blech-Hermoni Y, Ladd AN. RNA binding proteins in the regulation of heart development. Int J Biochem Cell Biol. 2013;45:2467–2478. doi: 10.1016/j.biocel.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Carreira-Rosario A, Bhargava V, Hillebrand J, Kollipara RK, Ramaswami M, Buszczak M. Repression of Pumilio protein expression by Rbfox1 promotes germ cell differentiation. Dev Cell. 2016;36:562–571. doi: 10.1016/j.devcel.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Wang YH, Chang MY, Lin CY, Weng CW, Westerfield M, Tsai HJ. Multiple upstream modules regulate zebrafish myf5 expression. BMC Dev Biol. 2007;3(7):1. doi: 10.1186/1471-213X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zubovic L, Yang F, Godin K, Pavelitz T, Castellanos J, Macchi P, Varani G. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 2016;44:4381–4395. doi: 10.1093/nar/gkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Richter JD. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol. 2012;4:a012344. doi: 10.1101/cshperspect.a012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen J, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Stoiber W, Hammond CL, Steinbacher P, Haslett JR, Barresi MJ, Patterson SE, Adiarte EG, Hughes SM. Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol Dev. 2006;8:101–110. doi: 10.1111/j.1525-142X.2006.05079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JE, Panavaite L, Gunther S, Wennekamp S, Groner AC, Pigge A, Salvenmoser S, Trono D, Hufnagel L, Hiiragi T. Venus trap in the mouse embryo reveals distinct molecular dynamics underlying specification of first embryonic lineages. EMBO Rep. 2015;16:1005–1021. doi: 10.15252/embr.201540162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Jensen KB. NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS One. 2011;6:e21585. doi: 10.1371/journal.pone.0021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont NA, Wang YX, Rudnicki MA. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development. 2015;142:1572–1581. doi: 10.1242/dev.114223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy S, Hargrave M, Knight R, Mebus K, Ingham PW. Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development. 2008;135:2115–2126. doi: 10.1242/dev.015719. [DOI] [PubMed] [Google Scholar]
- Feng X, Adiarte EG, Devoto SH. Hedgehog acts directly on the zebrafish dermomyotome to promote myogenic differentiation. Dev Biol. 2006;300:736–746. doi: 10.1016/j.ydbio.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol Cell. 2009;36:1007–1017. doi: 10.1016/j.molcel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, Conboy JG. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol. 2011;359:251–261. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Meera P, Stoilov P, Shiue L, O’Brien JE, Meisler MH, Ares M, Jr, Otis TS, Black DL. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurtzen K, Knopf F, Wehner D, Huitema LF, Schulte-Merker S, Weidinger G. Mature osteoblasts dedifferentiate in response to traumatic bone injury in the zebrafish fin and skull. Development. 2014;141:2225–2234. doi: 10.1242/dev.105817. [DOI] [PubMed] [Google Scholar]
- Gunther S, Kim J, Kostin S, Lepper C, Fan CM, Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013:S1934–5909. doi: 10.1016/j.stem.2013.07.016. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich DB, Nguyen PD, Siegel AL, Ehrlich OV, Sonntag C, Phan JM, Berger S, Ratnayake D, Hersey L, Berger J, Verkade H, Hall TE, Currie PD. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science. 2016;353:aad9969. doi: 10.1126/science.aad9969. [DOI] [PubMed] [Google Scholar]
- Hamada N, Ito H, Iwamoto I, Mizuno M, Morishita R, Inaguma Y, Kawamoto S, Tabata H, Nagata K. Biochemical and morphological characterization of A2BP1 in neuronal tissue. J Neurosci Res. 2013;91:1303–1311. doi: 10.1002/jnr.23266. [DOI] [PubMed] [Google Scholar]
- Hamada N, Ito H, Iwamoto I, Morishita R, Tabata H, Nagata K. Role of the cytoplasmic isoform of RBFOX1/A2BP1 in establishing the architecture of the developing cerebral cortex. Mol Autism. 2015;6:56. doi: 10.1186/s13229-015-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Ito H, Nishijo T, Iwamoto I, Morishita R, Tabata H, Momiyama T, Nagata K. Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci Rep. 2016;6:30805. doi: 10.1038/srep30805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Hollway GE, Bryson-Richardson RJ, Berger S, Cole NJ, Hall TE, Currie PD. Whole-somite rotation generates muscle progenitor cell compartments in the developing zebrafish embryo. Dev Cell. 2007;12:207–219. doi: 10.1016/j.devcel.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg R, Liu S, Blackburn JS, Linardic CM, Rosenberg AE, Nielsen PG, Mempel TR, Langenau DM. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell. 2012;21:680–693. doi: 10.1016/j.ccr.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson HE, Ono Y, Wang X, Elworthy S, Cunliffe VT, Ingham PW. The role of Sox6 in zebrafish muscle fiber type specification. Skelet Muscle. 2015;5:2. doi: 10.1186/s13395-014-0026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. Embo J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisúa Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B, Chong SW, He J, Wang X, Xu Y, Wan H, Tong Y, Yan T, Korzh V, Gong Z. Recapitulation of fast skeletal muscle development in zebrafish by transgenic expression of GFP under the mylz2 promoter. Dev Dyn. 2003;227:14–26. doi: 10.1002/dvdy.10273. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Glacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S. Pax3/7 mark a novel population of primitive myogenic cells during development. Genes & Development. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl TR, Shibata H, Vo T, Huynh DP, Pulst SM. Identification and expression of a mouse ortholog of A2BP1. Mamm Genome. 2001;12:595–601. doi: 10.1007/s00335-001-2056-4. [DOI] [PubMed] [Google Scholar]
- Knappe S, Zammit PS, Knight RD. A population of Pax7-expressing muscle progenitor cells show differential responses to muscle injury dependent on developmental stage and injury extent. Front Aging Neurosci. 2015;7:161. doi: 10.3389/fnagi.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, Weidinger G. Bone regeneration via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, Geschwind DH, Black DL, Martin KC. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Chen HY, Li YW, Wu SY, Wangta-Liu, Lin GH, Hu SY, Chang ZK, Gong HY, Liao CH, Chiang KY, Huang CW, Wu JL. Progranulin regulates zebrafish muscle growth and regeneration through maintaining the pool of myogenic progenitor cells. Science Rep. 2013;3:1176. doi: 10.1038/srep01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, Massirer KB, Pratt GA, Black DL, Gray JW, Conboy JG, Yeo GW. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- März M, Schmidt R, Rastegar S, Strähle U. Regenerative response following stab injury in the adult zebrafish telencephalon. Dev Dyn. 2011;240:2221–2231. doi: 10.1002/dvdy.22710. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, L’honore A, Buckingham M. Lying low but ready for action: the quiescent muscle satellite cell. FEBS J. 2013;280:4036–4050. doi: 10.1111/febs.12372. [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord H, Burguiere A, Muck J, Nord C, Ahlgren U, von Hofsten J. Differentiation regulation of myosin heavy chains defines new muscle domains in zebrafish. Mol Biol Cell. 2014;25:1384–1395. doi: 10.1091/mbc.E13-08-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord H, Dennhag N, Muck J, von Hofsten J. Pax7 is required for establishment of the xanthophore lineage in zebrafish embryos. Mol Biol Cell. 2016;27:1853–1862. doi: 10.1091/mbc.E15-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord H, Nygard Skalman L, von Hofsten J. Six1 regulates proliferation of Pax7-positive muscle progenitors in zebrafish. J Cell Sci. 2013;126:1868–1880. doi: 10.1242/jcs.119917. [DOI] [PubMed] [Google Scholar]
- O’Brien JE, Drews VL, Jones JM, Dugas JC, Barres BA, Meisler MH. Rbfox proteins regulate alternative splicing of neuronal sodium channel SCN8A. Mol Cell Neurosci. 2012;49:120–126. doi: 10.1016/j.mcn.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten C, van der Ven PF, Lewrenz I, Paul S, Steinhagen A, Busch-Nentwich E, Eichhorst J, Wiesner B, Stemple D, Strahle U, Furst DO, Abdelilah-Seyfried S. Xirp proteins mark injured skeletal muscle in zebrafish. PLoS One. 2012;7:e31041. doi: 10.1371/journal.pone.0031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti S, Giudice J, Dagnino-Acosta A, Knoblauch M, Singh RK, Hanna A, Mo Q, Hicks J, Hamilton S, Cooper TA. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function. Human Molecular Genetics. 2015;24:2360–2374. doi: 10.1093/hmg/ddv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipalia TG, Koth J, Roy SD, Hammond CL, Kawakami K, Hughes SM. Cellular dynamics of regeneration reveals role of two distinct Pax7 stem cell populations in larval zebrafish muscle repair. Dis Model Mech. 2016;9:671–684. doi: 10.1242/dmm.022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, Conboy JG. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16*. Journal of Biological Chemistry. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signaling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer MM, Sörensen I, Kuscha V, Frank RE, Liu C, Becker CG, Becker T. Motor neuron regeneration in adult zebrafish. J Neurosci. 2008;28:8510–8516. doi: 10.1523/JNEUROSCI.1189-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Rowlerson A, Radaelli G, Mascarello F, Veggetti A. Regeneration of skeletal muscle in two teleost fish: Sparus aurata and Brachydanio rerio. Cell Tissue Res. 1997;289:311–322. doi: 10.1007/s004410050878. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- Runfola V, Sebastian S, Dilworth FJ, Gabellini D. Rbfox proteins regulate tissue-specific alternative splicing of Mef2D required for muscle differentiation. J Cell Sci. 2015;128:631–637. doi: 10.1242/jcs.161059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saera-Vila A, Kasprick DS, Junttila TL, Grzegorski SJ, Louie KW, Chiari EF, Kish PE, Kahana A. Myocyte dedifferentiation drives extraocular muscle regeneration in adult zebrafish. Invest Ophthalmol Vis Sci. 2015;56:4977–4993. doi: 10.1167/iovs.14-16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saera-Vila A, Kish PE, Kahana A. Fgf regulates dedifferentiation during skeletal muscle regeneration in adult zebrafish. Cell Signal. 2016;28:1196–1204. doi: 10.1016/j.cellsig.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Seger C, Hargrave M, Wang X, Chai RJ, Elworthy S, Ingham PW. Analysis of Pax7 expressing myogenic cells in zebrafish muscle development, injury, and models of disease. Dev Dyn. 2011;240:2440–2451. doi: 10.1002/dvdy.22745. [DOI] [PubMed] [Google Scholar]
- Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: Defining the myogenic potency of aging skeletal muscle. Dev Biol. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Wleklinski-Lee M, Yablonka-Reuveni Z. Skeletal muscle satellite cells can spontaneously enter an alternative mesenchymal pathway. J Cell Sci. 2004;117:5393–5404. doi: 10.1242/jcs.01419. [DOI] [PubMed] [Google Scholar]
- Siegel AL, Gurevich DB, Currie PD. A myogenic precursor cell that could contribute to regeneration in zebrafish and its similarity to the satellite cell. FEBS J. 2013;280:4074–4088. doi: 10.1111/febs.12300. [DOI] [PubMed] [Google Scholar]
- Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simões M, Leon J, Roehl H, Cancela ML, Jacinto A. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- Stellabotte F, Dobbs-McAuliffe B, Fernandez DA, Feng X, Devoto SH. Dynamic somite cell rearrangements lead to distinct waves of myotome growth. Development. 2007;134:1253–1257. doi: 10.1242/dev.000067. [DOI] [PubMed] [Google Scholar]
- Stewart S, Stankunas K. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol. 2012;365:339–349. doi: 10.1016/j.ydbio.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11:583–585. doi: 10.1089/zeb.2014.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J, Maves L. Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip Rev Dev Biol. 2016;5:518–534. doi: 10.1002/wdev.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Moore JC, Ignatius MS, Tenente IM, Hayes MN, Garcia EG, Torres Yordán N, Bourque C, He S, Blackburn JS, Look AT, Houvras Y, Langenau DM. Imaging tumour cell heterogeneity following cell transplantation into optically clear immunedeficient zebrafish. Nat Commun. 2016;7:10358. doi: 10.1038/ncomms10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee JM, Sartori da Silva MA, Rygiel AM, Muncan V, Bink R, van den Brink GR, van Tijn P, Zivkovic D, Kodach LL, Guardavaccaro D, Diks SH, Peppelenbosch MP. asb11 is a regulator of embryonic and adult regenerative myogenesis. Stem Cells Dev. 2012;21:3091–3103. doi: 10.1089/scd.2012.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh le A, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, Chow E, Gonzales C, Leung HY, Solomon I, Bronner-Fraser M, Megason SG, Fraser SE. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 2011;25:2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raamsdonk W, Tekronnie G, Pool CW, van de Laarse W. An immune histochemical and enzymic characterization of the muscle fibres in myotomal muscle of the teleost Brachydanio rerio, Hamilton-Buchanan. Acta histochem. 1980;67:200–216. doi: 10.1016/S0065-1281(80)80024-9. [DOI] [PubMed] [Google Scholar]
- Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, Abou Elela S, Roux P, Lemaitre JM, Tazi J. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013;4:2480. doi: 10.1038/ncomms3480. [DOI] [PubMed] [Google Scholar]
- von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci USA. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Wang QJ, Wang C, Reinholt B, Grant AL, Gerrard DE, Kuang S. Heterogeneous activation of a slow myosin gene in proliferating myoblasts and differentiated single myofibers. Dev Biol. 2015;402:72–80. doi: 10.1016/j.ydbio.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman RE. Development of the lateral musculature in the teleost, Brachydanio rerio: A fine structural study. Am J Anat. 1969;125:457–493. doi: 10.1002/aja.1001250406. [DOI] [PubMed] [Google Scholar]
- Wei C, Qiu J, Zhou Y, Hu J, Ouyang K, Banerjee I, Zhang C, Chen B, Li H, Chen J, Song LS, Fu XD. Repression of the Central Splicing Regulator RBFox2 is functionally linked to pressure overload-induced heart failure. Cell Rep. 2015;10:1521–1533. doi: 10.1016/j.celrep.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, Krainer AR, Darnell RB, Zhang C. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windner SE, Doris RA, Ferguson CM, Nelson AC, Valentin G, Tan H, Oates AC, Wardle FC, Devoto SH. Tbx6, Mesp-b and Ripply1 regulate the onset of skeletal myogenesis in zebrafish. Development. 2015;142:1159–1168. doi: 10.1242/dev.113431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Huang SC, Wu JY, Benz EJ., Jr Regulated Fox-2 isoform expression mediates protein 4.1R splicing during erythroid differentiation. Blood. 2008;111:392–401. doi: 10.1182/blood-2007-01-068940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- Zhang H, Anderson JE. Satellite cell activation and population on single muscle-fiber cultures from adult zebrafish (Danio rerio) J Exp Biol. 2014;217:1910–1917. doi: 10.1242/jeb.102210. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hasman RA, Barron VA, Luo G, Lou H. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol Biol Cell. 2006;17:5105–5114. doi: 10.1091/mbc.E06-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


