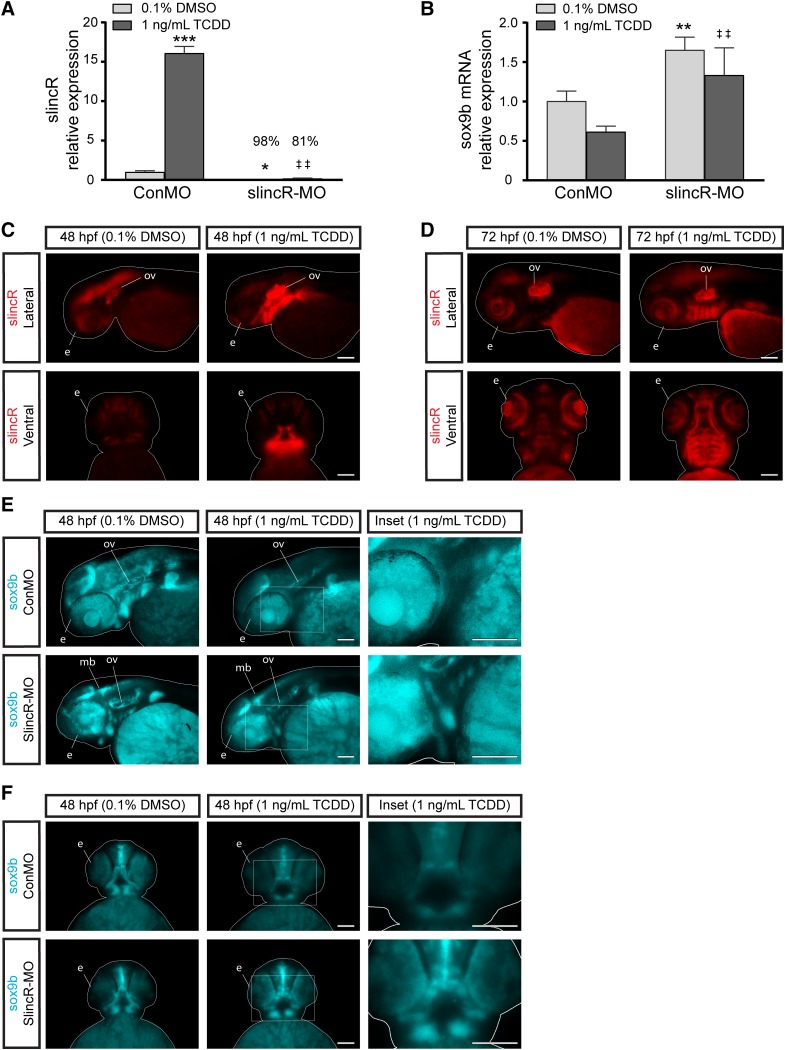
Fig. 4.
Comparative analysis of the relative expression of slincR and sox9 in TCDD-exposed samples. SlincR (A) and sox9b mRNA (B) quantitative expression in 48 hpf whole embryo slincR (slincR-MO) and control (ConMO) morphants developmentally exposed to 0.1% DMSO (vehicle control) or 1 ng/ml TCDD. For all quantitative PCR data, expression values were analyzed using the 2−ΔΔCT method. Expression values were normalized to β-actin and the control morphants served as the calibrator. Samples represent a minimum of three biologic replicates. Results were statistically analyzed using the GraphPad Prism 7.02 software. The data were tested for normality using the Shapiro-Wilk normality test and analyzed with two-way analysis of variance and correction for multiple comparisons was performed using the Dunnett’s test (95% confidence intervals). Error bars indicate S.D. of the mean; *P < 0.05, **P < 0.01, ***P < 0.001 compared with controls; ##P < 0.01 compared with TCDD control. (C–F) dorsal and lateral views of in situ hybridization samples targeting slincR (red) and sox9b (blue) in 48 hpf (C, E, and F) and 72 hpf (D) embryos (n = 6–10 embryos). (C and D) Representative images from control morphants. In the TCDD-exposed samples, slincR morphants displayed increased sox9b expression patterns compared with controls (white rectangle, which represents the magnified image in the inset). The fish are outlined with a white line; e = eye, ov = otic vesicle, p = pectoral fin, and mb = midbrain; 100 µm scale bar. All experiments were independently repeated a minimum of two times.