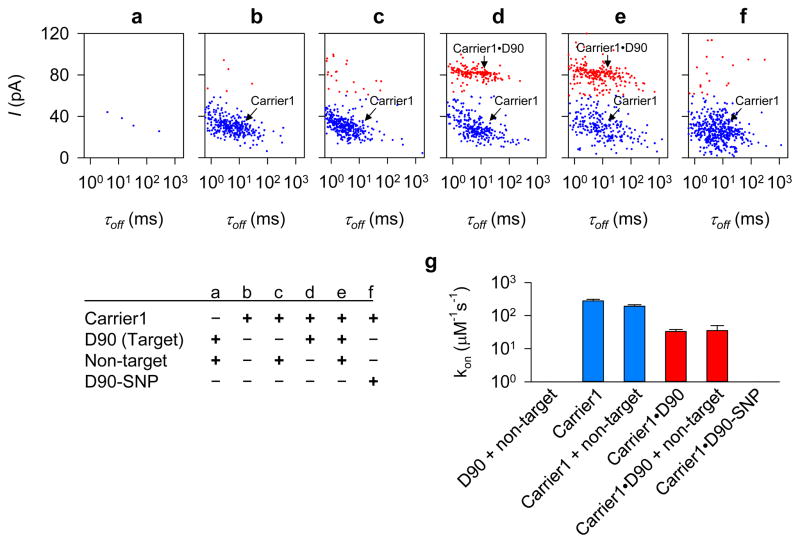
Figure 4.
Selective and interference-free detection of target gene fragment in the presence of non-target species. a–f, Scatter plots of blockade current amplitude (I) and duration (τoff) for the mixture of D90 (target) and non-target DNA (a), free Carrier1 (b), Carrier1 in the presence of non-target DNA (c), the mixture of Carrier1 and D90 in the absence (d) and presence (e) of non-target DNA, and the mixture of Carrier1 and D90-SNP. Blue and red dots indicate the blockades associated with the capture of free carrier and carrier•DNA complexes, respectively. Non-target DNA mixture included pT7 plasmid DNA (10.2 ng·μL−1) and a 225-bp BRAF gene fragment PCR reaction mixture (37.2 ng·μL−1). D90-SNP is a variant of D90, which forms a single mismatched basepair with Carrier1 (Table S1, Fig. S14 for traces). g, Comparison of the capture rate (kon) for the carrier•DNA complex for conditions shown in a–f. The presence of non-target DNA did not influence the capture efficiencies for both free nanocarrier and the carrier•DNA complex, suggesting that the nanocarrier is specific to the target and does not bind to non-target DNA, and that the formation of the carrier•DNA complex is not influenced by non-target DNA.