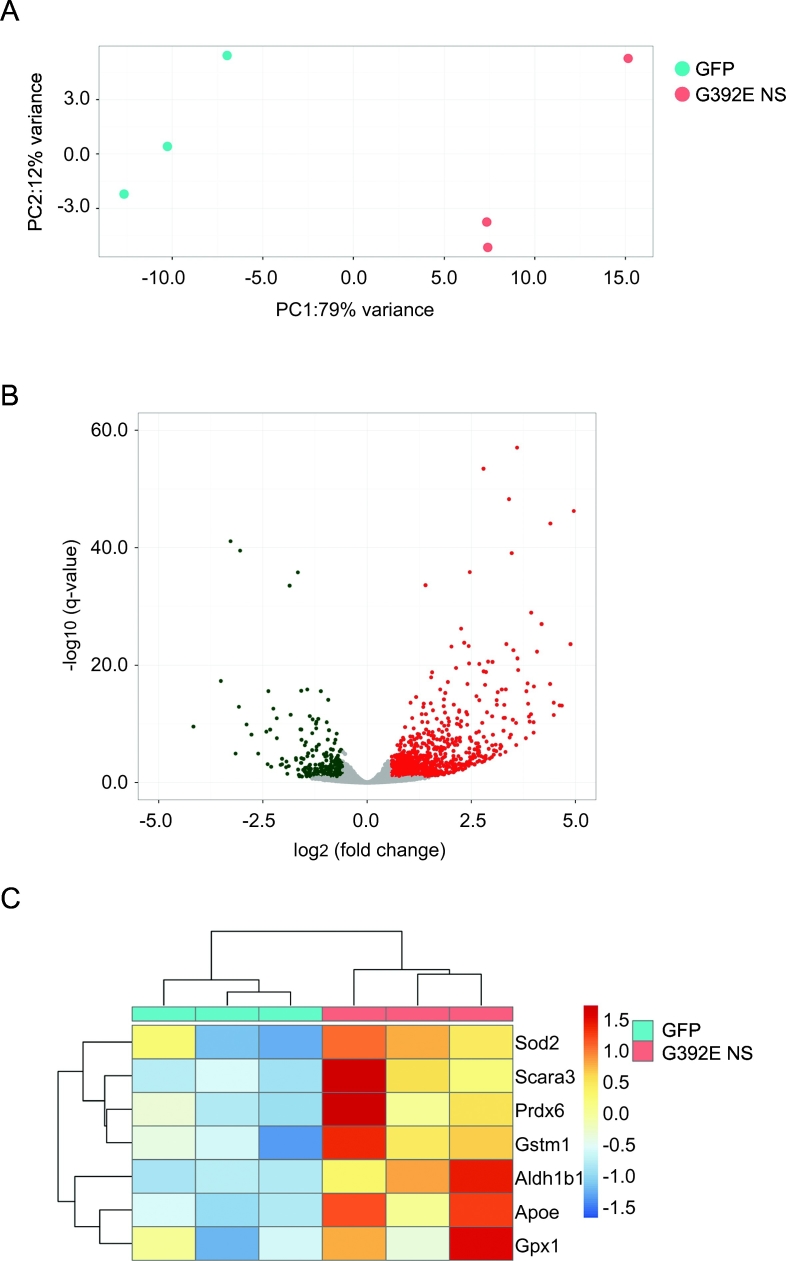
Fig. 4.
RNA sequencing analysis of differentiated NPCs overexpressing GFP and G392E NS. NPCs stably expressing GFP or G392E NS were differentiated for 6 days in vitro, and their total RNA recovered and analysed by next generation sequencing. A. Results of principal component analysis on RNA sequencing data from three GFP (red dots) and three G392E NS (blue dots) independent samples. The plot shows the first two principal components (PC1, PC2). GFP and G392E samples showed high similarity between replicates and clustered according to their respective genotypes. B. Volcano plot of log2 fold-change (x-axis) versus − log10 q-value (y-axis, representing the probability that the gene is differentially expressed) of the results of RNA sequencing analysis of GFP and G392E NS samples. 1960 genes were significantly downregulated (green dots) or upregulated (red dots) in G392E NS cells in comparison with GFP cells. C. Heat-map of the regularised-logarithm transformed scaled counts (Love et al., 2014) of the results of RNA sequencing analysis for the indicated genes involved in oxidative metabolism. Scara3, Prdx6, Gstm1, Aldh1b1 and Apoe showed consistent upregulation in G392E NS samples, while higher variability across experimental replicates was detectable for Sod2 and Gpx1. Condition tree (top) and gene tree (left side) are represented.