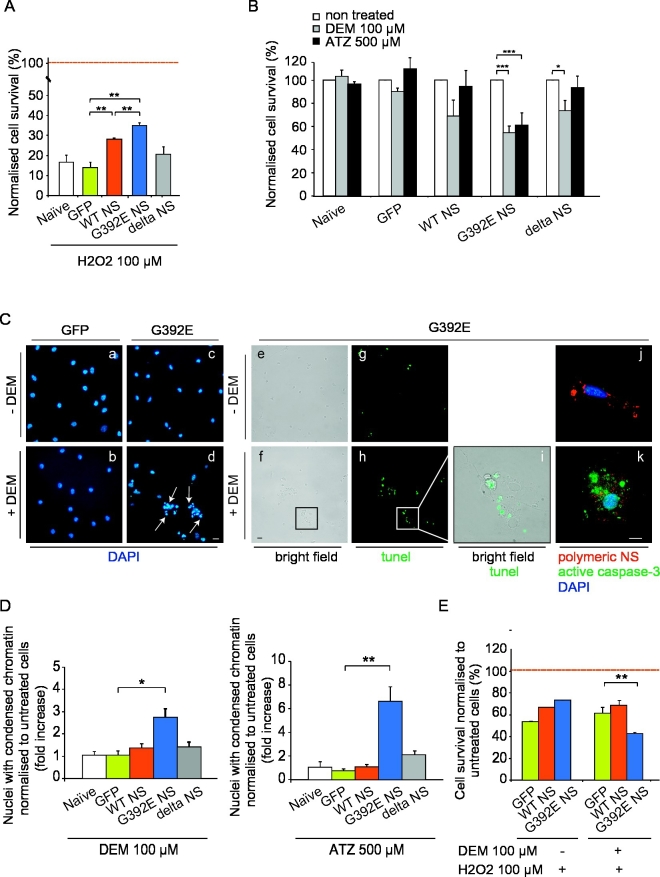
Fig. 6.
Polymers of G392E NS cause oxidative stress in differentiated NPCs. A. NPCs were differentiated for 6 days and each cell line was incubated in the absence or presence of H2O2 (100 μM) for 30 min. Cell survival was quantified 24 h later by counting DAPI-positive cells. Results are expressed as the percentage of surviving neurons compared with control cultures not treated with H2O2 (100% survival reference line), and are the mean +/− SEM of three independent repeats, t-test: **p ≤ 0.01. B. Differentiated NPCs were incubated for 1 h in control conditions (non treated) or with either the glutathione depletor DEM (100 μM) or the catalase inhibitor ATZ (500 μM). Neuronal survival was quantified 24 h later by counting DAPI-positive stained cells. Results are expressed as the percentage of surviving neurons compared to control cultures, and are the mean +/− SEM of three independent repeats, t-test: *p ≤ 0.05; ***p ≤ 0.005. C. Differentiated NPCs expressing GFP or G392E NS were treated or not with 100 μM DEM and stained with DAPI (a–d), TUNEL assay (e–i) or anti-cleaved caspase-3 antibody (green) and anti-NS polymers (7C6 antibody, red) (j–k), to assess the nature of nuclei showing chromatin condensation. Arrows: nuclei with condensed chromatin. D. Differentiated cells were incubated for 1 h with either 100 μM DEM or 500 μM ATZ and their nuclear morphology was analysed 24 h later. The toxic effect of DEM and ATZ was quantified by counting DAPI-stained nuclei with condensed chromatin. Results are expressed as the ratio between treated and non-treated cells, and data are expressed as the mean +/− SEM of three independent repeats, t-test: *p ≤ 0.05; **p ≤ 0.01. E. Differentiated NPCs were pre-incubated for 1 h without or with 100 μM DEM and then further incubated for 30 min with 100 μM H2O2 in the presence or absence of DEM. Neuronal survival was quantified 24 h later by counting DAPI-stained cells. Results are expressed as the percentage of surviving neurons compared with control non-treated cultures, and data are the mean +/− SEM of three independent repeats, t-test: **p ≤ 0.01. In panels A, B, D and E, 15 different fields (to a total of 150 to 350 cells) were counted per condition in each independent repeat.