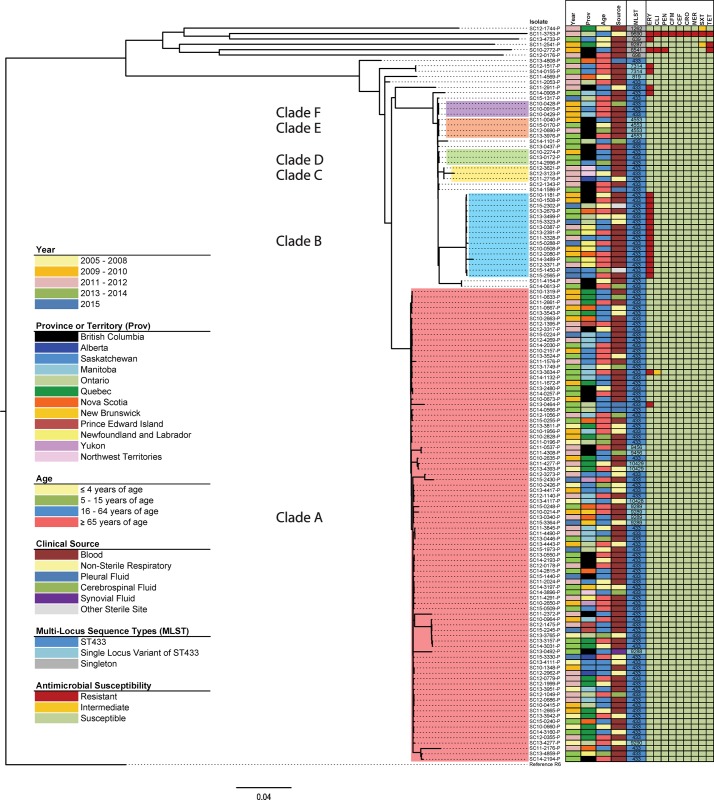
Fig 3. Whole genome core SNV maximum likelihood phylogenetic tree of 137 Streptococcus pneumoniae serotype 22F isolates collected in Canada from 2005–2015.
The maximum likelihood phylogenetic tree is rooted on the reference genome of S. pneumoniae R6 (GenBank accession no. NC_003098.1) and the scale bar represents the estimated evolutionary divergence between isolates on the basis of average genetic distance between strains (estimated number of substitutions in the sample / total number of high quality SNVs). Clades A through F identified by cluster analysis are denoted with shading. Coloured columns in the right side heatmap represent: year of isolation (Year); province or territory isolated (Prov); patient age group (Age); clinical isolation source (Source); multi-locus sequence type (MLST); and antimicrobial susceptibilities to erythromycin (ERY), clindamycin (CLI), penicillin (PEN), cefepime (CFM), cefotaxime (CEF), ceftriaxone (CRO), meropenem (MER), trimethoprim/sulfamethoxazole (SXT) and tetracycline (TET).