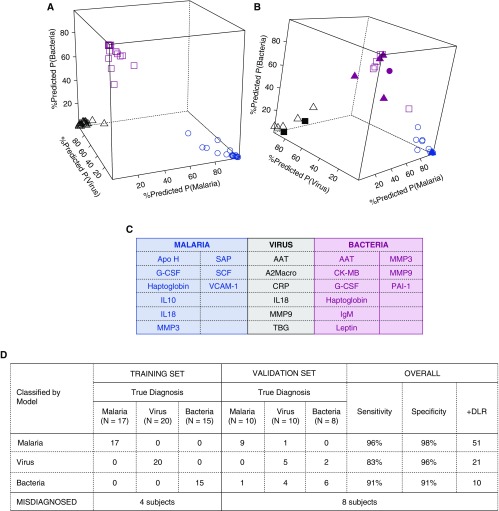
Figure 5.
Biomarker signature for febrile clinical pneumonia selected by multinomial logistic regression models (fitted via elastic net penalty). Predicted probabilities of diagnoses of malaria, virus, or bacteria in the training set (A) in which all patients were correctly classified, and in the validation set (B), in which some patients were misclassified. Blue, black, and pink represent malaria-, virus-, and bacteria-assigned diagnosis, respectively. Open shapes represent patients with matched true and assigned diagnosis (circles, malaria; triangles, virus; squares, bacteria). Solid shapes represent patients misclassified; for example, solid black squares are patients who were assigned a diagnosis of virus but were true bacteria patients, solid pink triangles are patients who were assigned a diagnosis of bacteria but were true virus patients, and solid pink circles are patients who were assigned a bacteria diagnosis but were true malaria patients. In C, markers selected in the support vector machine biomarker signature are listed and in D, training and validation errors. Overall, this signature was an excellent classifier for malaria, good for bacteria, and poorer classifier for virus patients. +DLR = positive diagnostic likelihood ratio; A2Macro = α2-macroglobulin; AAT = alpha-1 antitrypsin; Apo H = apolipoprotein H; CK-MB = creatine kinase–MB; CRP = C-reactive protein; G-CSF = granulocyte colony-stimulating factor; MMP3 = matrix metalloproteinase 3; MMP9 = matrix metalloproteinase 9; PAI-1 = plasminogen activator inhibitor 1; SAP = serum amyloid P-component; SCF = stem cell factor; TBG = thyroxine-binding globulin; VCAM-1 = vascular cell adhesion molecule 1.