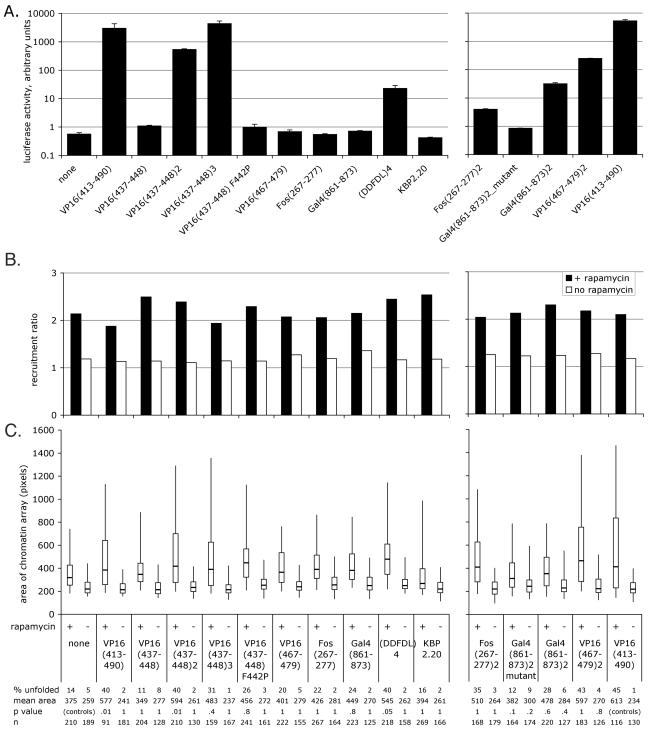
FIG. 4.
Transcriptional activity and large-scale chromatin decondensation activity of acidic activator motifs, all fused to FRB*-YFP. none, FRB*-YFP with no protein fused to it. See the legend to Fig. 3 for details. Error bars show SEMs. (A) Transient transcription assay. (B) Recruitment to the chromatin array. Sample labels are the same as for panels A and C. (C) Chromatin-unfolding assay. The initial experiment is shown. A subsequent experiment yielded similar results, except that the area of Gal4(861-873)2 was larger in the presence of rapamycin in the second experiment, and the area of VP16(437-448) was smaller in the absence of rapamycin and larger in the presence of rapamycin in the second experiment than in the first. The samples in the left and right charts were prepared separately and therefore statistically analyzed separately; note that the P values for the right side are compared to those of the positive control. The samples shown on the left (B and C) were prepared and analyzed at the same time as the samples in Fig. 3, so “none” and “VP16(413-490)” are the same data sets as shown in Fig. 3B and C.