Figure 1.
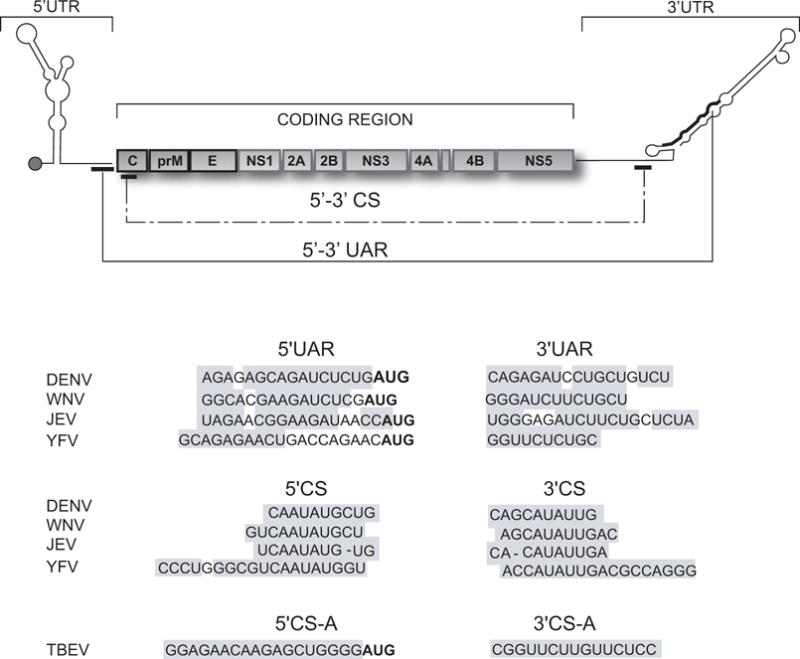
Flavivirus genome. On the top, schematic representation showing the coding and untranslated regions (UTRs) of the viral genome. The three structural proteins C, capsid; prM precursor to membrane protein; E, envelope; and the seven non-structural (NS) proteins NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 are indicated. The location of the complementary sequences 5′-3′UAR and 5′-3′CS of mosquito-borne flaviviruses is also shown. The bottom panels show the nucleotide sequences of the complementary regions 5′-3′CS and 5′-3′UAR of dengue virus type 2 (DENV2, Genebank number U87412), West Nile virus (WNV, Genebank number M12294), Japanese encephalitis virus (JEV, Genebank number NC001437), and yellow fever virus (YFV, Genebank number NC002031); and the 5′-3′CSA of tick-borne encephalitis virus (TBEV, Genebank number U27495). The grey boxes denote the inverted complementary sequences. In boldface the translation initiator AUG is indicated.
