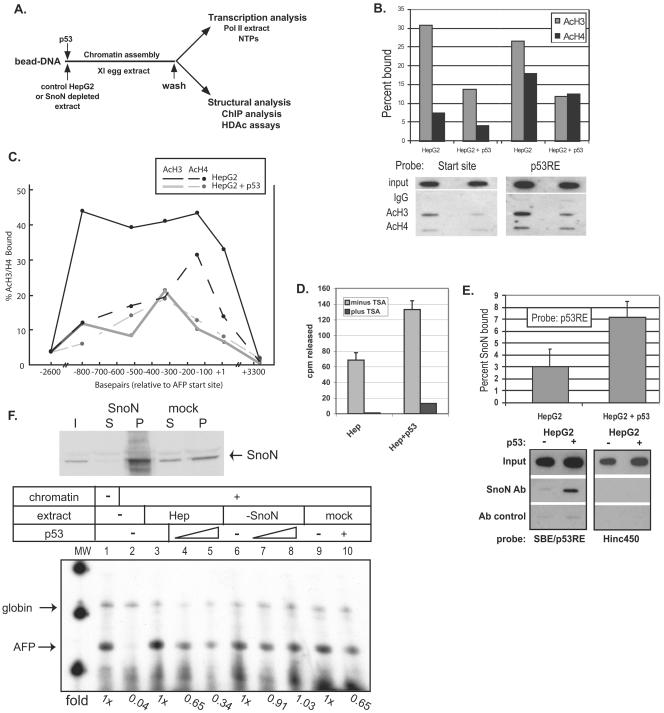
FIG. 6.
p53 targets histone deacetylation to the SBE/p53RE and mediates SnoN-dependent repression of AFP transcription. (A) Diagram of in vitro chromatin assembly and analysis. AFP bead DNA is preincubated with protein extracts and/or recombinant p53 protein prior to chromatin assembly over a 1-h incubation in X. laevis (Xl) egg extract. Assembled chromatin bead DNA is then washed in an isotonic buffer prior to functional assay by transcription in HeLa extract or structural analysis by ChIP and HDAC assays. (B and C) p53 triggers deacetylation of local and distal chromatin. ChIP assays (two to three independent assays for each data point) were performed in vitro on AFP/lacZ DNA, which was chromatin assembled in the presence of HepG2 whole-cell extract or HepG2 plus 500 ng of activated p53. DNA was purified from protein-chromatin immunoprecipitates and analyzed for percent SBE/p53RE and transcription start site DNA bound to acetylated histone H3 (AcH3) or histone H4 (AcH4) by Southern blotting with labeled (B) SBE/p53RE or start site (+4 to +26) oligonucleotides or (C) across the AFP templates with oligonucleotides specific for +10/+32, −140/−118, −327/−305, −520/−398, −800/−778, −2637/−2615, and +3339/+3361. Representative data are shown below the summary graph (B). (D) p53-dependent HDAC activity at SBE/p53RE. Immobilized SBE/p53RE DNA oligomers were incubated in 100 μg of HepG2 extract, either tryptic soy agar treated (10 μM; black bars) or untreated (gray bars), in the presence or absence of recombinant, activated p53. Protein complexes bound to the oligomers were washed and incubated with in vivo-labeled, 3H-acetylated histones, which were isolated and purified. HDAC enzymatic activity was measured by scintillation counting of released [3H]acetyl groups. The graphrepresents average values from multiple experiments and standard deviations of the means. (E) p53-dependent SnoN association at the SBE/p53RE. A series of ChIP studies (≥3 for each data point) were performed as described above. Values for percentage of SnoN bound relative to input at the SBE/p53RE in the presence and absence of recombinant p53 were normalized to antibody (Ab) control reactions. Probes used for Southern blots of precipitated DNA were labeled SBE/p53RE oligonucleotides or Hinc450 oligonucleotides (+3333 to +3356), which will detect nonspecific binding to AFP/lacZ DNA. (F) SnoN is required for p53-mediated repression of in vitro AFP chromatin transcription. HepG2 whole-cell extract was immunodepleted for SnoN protein (upper panel) and then analyzed by in vitro chromatin transcription (lower panel). Upper panel: 40 μg of protein of starting HepG2 extract, input (I), equal levels of protein from depleted extract, supernatant (S) and pellet (P) derived from boiled antibody-coupled beads used for immunodepletion. Lower panel: immobilized AFP DNA templates were incubated in (i) control (lanes 3 to 5; approximately 200 μg of total protein), (ii) immunodepleted (lanes 6 to 8; approximately 200 μg of total protein), or (iii) mock-depleted (lanes 9 and 10; approximately 200 μg of total protein) extracts or (iv) buffer (lanes 1 and 2) in the presence of recombinant p53 (500 ng, lanes 4 and 7; 1 μg, lanes 5, 8, and 10) or p53 dialysis buffer (lanes 1 to 3, 6 and 9) prior to chromatin assembly. Chromatin-assembled DNA templates were washed in nuclear extract buffer and in vitro transcribed in HeLa nuclear extract. Chick β-globin (50 ng) DNA was added during the transcription period as an RNA recovery control (57). AFP and Globin primer extension products are indicated. n-fold changes of transcribed AFP in vitro RNA signals are shown below each data point.