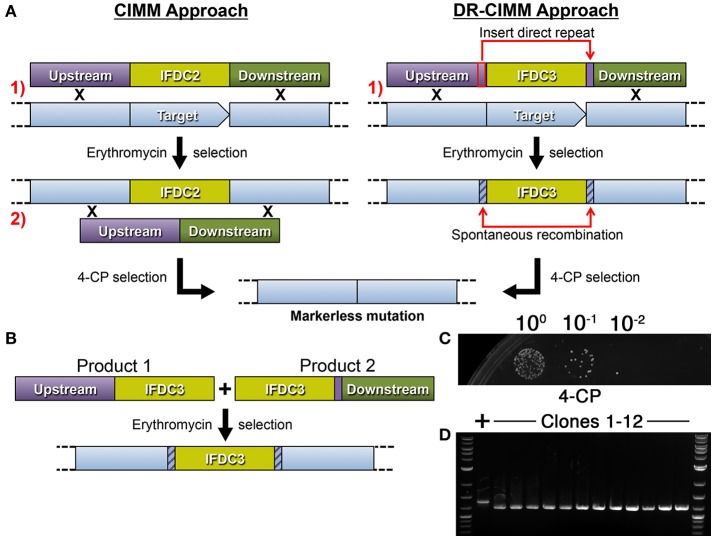
Figure 2.
Direct repeat-mediated cloning-independent markerless mutagenesis (DR-CIMM). (A) Comparison of the CIMM and DR-CIMM approaches. Individual transformation steps are numbered in red. The previous IFDC2-based CIMM approach (shown on the left) requires two sequential transformations with two separate OE-PCR products. The first transformation inserts the IFDC2 cassette onto the chromosome via double crossover homologous recombination and transformants are selected based upon acquired erythromycin resistance. The second transformation excises the IFDC2 cassette to render the cells 4-CP-resistant. The current IFDC3-based DR-CIMM approach (shown on the right) requires a single transformation with an OE-PCR product. The mutagenesis construct contains a small segment of the 3' end of the upstream homologous fragment (outlined in red) inserted immediately after the IFDC3 cassette, which serves as the direct repeat. After transformation and selection on erythromycin, clones are cultured in non-selective medium to excise the cassette due to spontaneous recombination between the two direct repeats. These clones are subsequently isolated due to their acquired 4-CP resistance. (B) Illustration of the two-fragment DR-CIMM construct assembly approach. To avoid interference from the direct repeats during OE-PCR assembly, two smaller segments of the final DR-CIMM construct are assembled by OE-PCR, separating the direct repeats between the two fragments. The first OE-PCR product is created by attaching the IFDC3 cassette onto the 3' of the upstream homologous fragment, while the second OE-PCR product is created by attaching the direct repeat and downstream homologous fragment onto the 3' of IFDC3 cassette. Thus, both OE-PCR products contain a copy of IFDC3. The two OE-PCR products are transformed simultaneously into S. mutans and selected for antibiotic resistance. Homologous recombination between the OE-PCR products assembles the final construct in vivo, which is then recombined with the chromosome. (C) A 156 bp deletion construct was created using IFDC3 and the previously described DR-CIMM methodology. Shown here are the results obtained from the final 4-CP negative selection step. (D) 12 CFU were randomly selected from the 4-CP plates and PCR-amplified to compare their genotypes. All clones exhibited the expected 156 bp deletion, resulting in smaller PCR amplicons relative to the parent wild-type (+).