FIGURE 6.
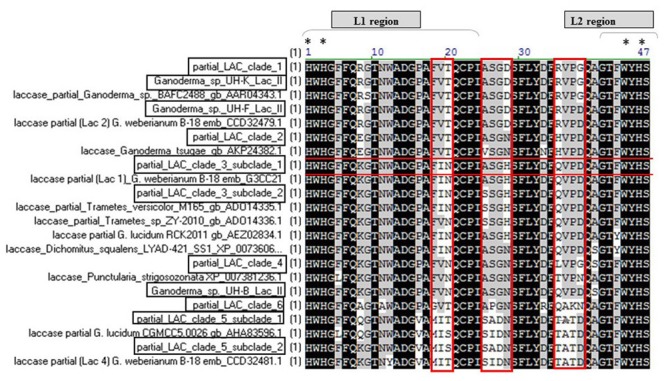
Extracted fragment from the multiple alignment the deduced amino acid sequences of laccase fragments from Ganoderma strains with the laccases presented in the phylogenetic tree. The position residues conserved in all sequences are highlighted in black. The positions conserved in 80 and 60% of the sequences, respectively, are highlighted in dark and light gray. The L1 region and a section of L2 region present in the studied fragments as well as the histidine residues involved in copper-binding (T2 and T3-sites ligands marked with asterisks) of the active site are indicated. LAC clade 1 (UH-I lac II, UH-A lac II, UH-H lac I, UH-K lac II, UH-C lac II, UH-M lac V, UH-G lac I, UH-E lac II), LAC clade 2 (UH-M lac IV, UH-B lac IV, UH-G lac III, UH-H lac II, UH-C lac IV, UH-A lac IV, UH-B lac III), LAC clade 3 subclade 1 (UH-C lac III, A lac III, G lac II), LAC clade 3 subclade 2 (UH-F lac III, UH-D Lac II), LAC clade 4 (UH-M lac II, UH-L lac I), LAC clade 5 subclade 1 (UH-J lac I, UH-C lac I, UH-D lac I, UH-F lac I, UH-K lac I, UH-I lac I, UH-M lac I, UH-E lac I), LAC clade 5 subclade 2 (UH-A lac I, UH-B lac I, UH-J lac II) LAC clade 6 (UH-I lac III, UH-M lac III, UH-L lac II).
