Fig. 4.
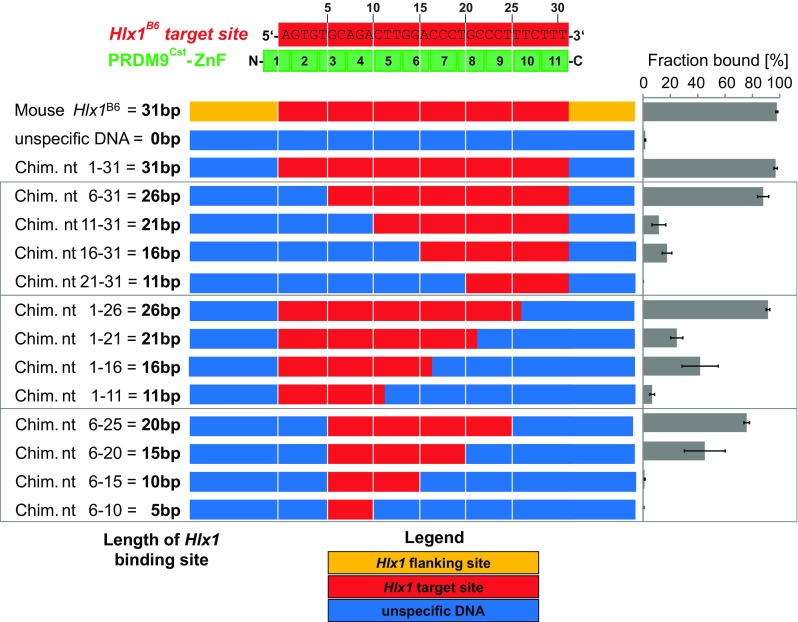
Binding specificity of the PRDM9 ZnF domain. Shown is the alignment of the Hlx1 B6 binding site and the PRDM9Cst-ZnF repeats. Each zinc finger contacts three nucleotides of the binding site. The numbers indicate the ZnFs’ position to the respective nucleotide sequence. The complex formation was assessed by incubating for 20 min 2.5 μM PRDM9Cst-ZnF (YFP tagged) with 15 nM of different arrangements of the Hlx1 B6 sequence (10 mM Tris-HCl pH 7.5, 50 mM KCl, 1 mM DTT, 50 ng/μl polydIdC, 0.05% NP-40, 50 μM ZnCl2) and evaluated by EMSA. The used DNA concentration was optimized to be much lower than the K D of the substrate of the highest affinity to ensure non-saturating conditions. The flanking sites (orange) were replaced by an unspecific DNA sequence (blue) to create the chimera fragment nt 1–31, which contains the 31 bp target site (red) of PRDM9Cst. Furthermore, this target site (red) was replaced in five-nucleotide steps by an unspecific DNA sequence (blue), as shown. The fraction bound was calculated as percentage of the complex (ratio of pixel intensities of the shifted band to the sum of free and bound DNA). The error bars represent the standard deviation of three independent experiments
