Fig. 5.
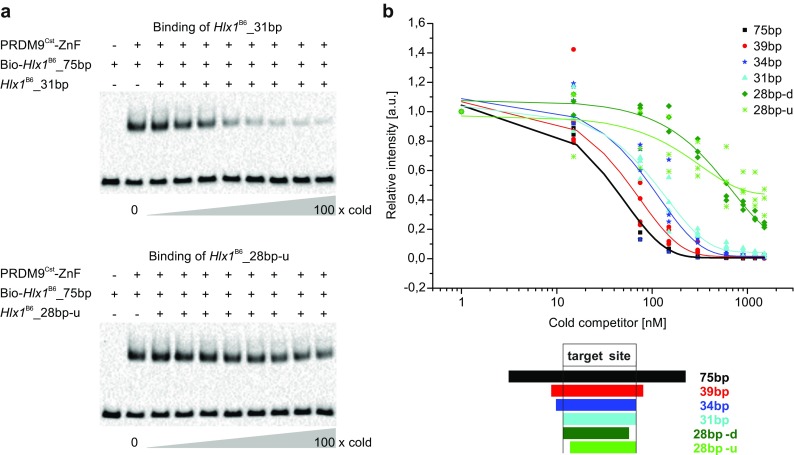
PRDM9 binding to different lengths of DNA. a In this competition EMSA, 250 nM PRDM9Cst-ZnF (YFP tagged) was incubated simultaneously with hot (biotinylated) reference DNA (75 bp Hlx1 B6) and different ratios of excess cold Hlx1 B6 for 1 h (a time frame that showed unchanged complex amounts; Supplementary_Fig_S4). We used for each experiment different lengths of cold DNA (75 bp, 39 bp, 34 bp, 31 bp, 28 bp-d, and 28 bp-u sequences are shown in Supplementary_Table_S1, section C) representing truncations of the 75 bp Hlx1 B6 sequence. Shown are representative EMSAs for the cold competitors with 31 and 28 bp. The measurements were performed in triplicates for each cold competitor. b Intensities of the shifted bands are calculated relative to the reference PRDM9Cst-Hlx1 B6 complex without the addition of cold DNA (lane 2) and are plotted as relative intensities against the concentration of the cold competitor in a semi-logarithmic graph. 28 bp-d and 28 bp-u indicate 28 base-pair long fragments with truncations of three bases from the target binding site at either the downstream (3′ end) or the upstream (5′ end) end, respectively
