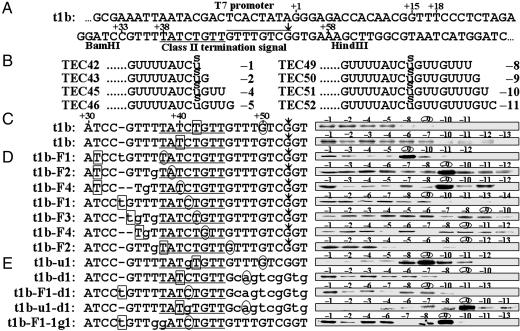
Fig. 2.
Photocross-linking of RNA and RNA polymerase. (A) The template t1b, containing the E. coli rrnB t1 class II termination signal, terminates T7 RNA polymerase transcription at +53 (with respect to the 5′ end of the RNA), indicated by an arrow. The CS module is underlined, and the T rich module overlined. (B) RNA sequences of t1b elongation complexes obtained by stepwise walking after an sU was incorporated into radiolabeled RNA at +42. The positions of sU with respect to the 3′ end of RNA are indicated in negative numbers. (C) The complexes in B were irradiated for photocross-linking (first line). Another series of t1b elongation complexes were obtained after an sU was incorporated into RNA at boxed position +40, followed by irradiation (second line). (D) Template variants t1b-F1 through -F4 differ from t1b only at the positions indicated by lowercase letters, and all are active in termination. Seven series of elongation complexes were obtained after sU was incorporated at the boxed positions in RNA. Complexes that will have an sU at -9 RNA residue are indicated in the sequences by the circled positions of their 3′ ends. (E) Five mutants differing from t1b at the positions indicated by lowercase letters. All are inactive in termination. The efficiencies of the “strong” cross-linking (in the six complexes mentioned in the text) averaged over five experiments range from 14% to 21% (an average of 17%), whereas those of the “weak” cross-linking are all <6.0%.