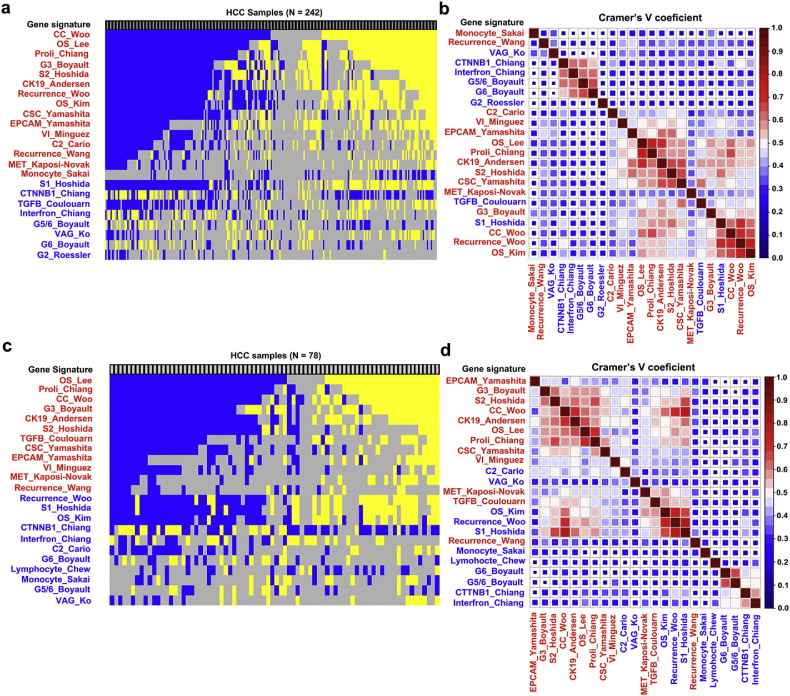
Fig. 1.
Nearest template prediction (NTP) results and their concordance with gene signatures in the GSE14520 and GSE54236 datasets. (a) NTP results in dataset GSE14520 (N = 242), with each column representing the prediction results for individual patients. Gene signatures suggesting poorer overall survival (OS) are labeled in blue, and signatures that suggest better OS are labeled in yellow (FDR, P < 0.05). The gray column indicates the presence of an unclassified group of patients (FDR, P > 0.05). The left label indicates the gene signature name, as listed in Table 1. (b) Heat map of Cramer's V coefficient values for pair-wise gene signatures in GSE14520; the signatures are clustered according to their degree of correlation. (c) NTP results in dataset GSE54236 (N = 78), with each column representing the prediction result for individual patients. Poorer (FDR, P < 0.05), better (FDR, P < 0.05), or unclassified (FDR > 0.05) outcomes for each patient are labeled in blue, yellow, or gray, respectively. The left label indicates the gene signature name as listed in Table 1. (d) Heat map of Cramer's V coefficient values for pair-wise gene signatures in GSE54236; the signatures are clustered according to their degree of correlation. Gene signatures that are significantly associated with the OS of HCC patients are labeled in red, and those not associated with OS are labeled in blue.