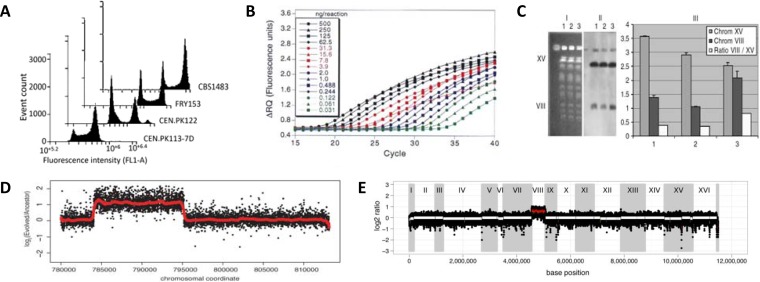
FIG 1.
Methods to analyze chromosome copy number and DNA content in yeast cells. (A) Absolute quantification of the DNA content of strain CBS1483 by flow cytometry using the DNA-intercalating dye Sytox Green and calibration with three strains of known ploidy. (Adapted from reference 23.) (B) qPCR fluorescence profiles for different initial concentrations of a template DNA sequence can be used to infer the amount of initial template in a reaction and to calculate relative copy numbers of different parts of the template DNA. (Republished from reference 157.) (C) Chromosome copy number determination of S. cerevisiae variants using contour-clamped homogeneous electric field electrophoresis and Southern blotting. I, stained CHEF gel; II, Southern blot hybridization; III, quantification of the hybridization bands. Lanes 1 and 2 show two disomic knockout strains that have only a single copy of chromosome VIII, while lane 3 shows a diploid control strain. (Modified from reference 28 with permission [copyright 2005 John Wiley & Sons Ltd.].) (D) Copy number estimation of chromosome II by array comparative genomic hybridization of an evolved strain relative to its unevolved parental strain. Deviating copy number can be detected by significant deviations of the measured signal and has been accentuated by a red line. (Republished from reference 108.) (E) Copy number estimation of the genome of the wine production strain VL3, based on whole-genome sequencing and read depth analysis. A marked increase of the read depth for chromosome VIII indicates a gain of copy of that chromosome. (Adapted from reference 117.)