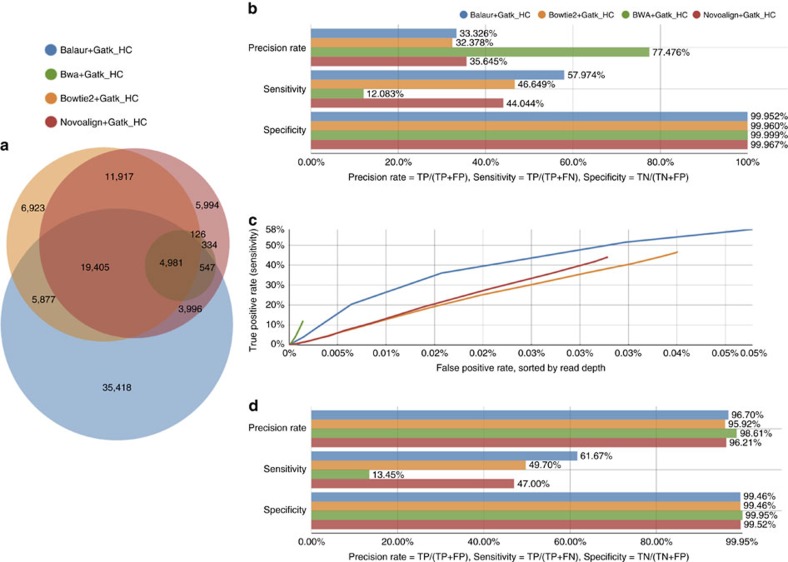
Figure 4. Evaluation of variant calling using the GCAT benchmarking platform.
Results are shown for the NA12878 Ion Torrent 225-bp SE exome 30 × data set available through GCAT22. Reads were aligned against the UCSC hg19 human genome reference. Balaur variant calling was performed using the GATK HaplotypeCaller pipeline23 (see Supplementary Note 2 for details). Comparisons are shown against the Bowtie2, BWA and Novoalign aligners with GATK HaplotypeCaller variant calling reports available through GCAT. All subfigures were generated by GCAT given the VCF file containing the Balaur variant calling results. (a) Venn diagram showing the concordance of the results obtained for each aligner. (b,c) Comparison against the NIST Genome in a Bottle SNP and indel call set (v2.18). (d) Comparison against the HumanOmni2.5-8v1 Illumina genotyping array. For more information about the benchmarks please see GCAT22.