Fig. 2.
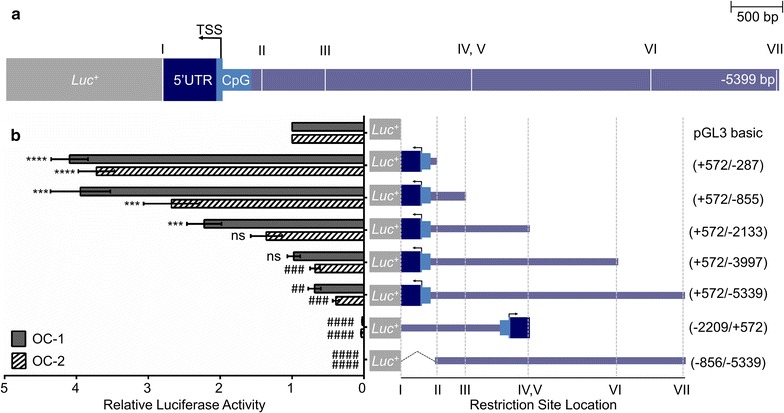
Promoter construct cartoon and luciferase assay. a αAtp2b2 promoter elements and restriction sites. The gene for Atp2b2 is located on the reverse strand oriented from right to left. This schematic represents the full promoter construct cloned into the luciferase vector. The full proximal promoter construct contains the three exons of the 5′ UTR (Iα, IIα, IIIα) and the 1st translated exon (Exon 1) up to the TSS (Table 4). Together these exons account for about 572 bases in the total construct. The rest of the 5.5 kbp construct contains the DNA directly upstream of the Iα exon (purple bar). The CpG island is contained between +62 and −219 bases around the TSS (blue bar). The restriction sites used to create promoter construct truncations are indicated by Roman numerals on the cartoon, their exact locations can be found in Table 1. b The luciferase assay indicates that the minimal promoter elements are contained in the 5′ UTR and CpG island. The promoter is directional. Transcription is not initiated without the CPG island and the 5′ UTR. Activator elements are contained within the first 1.5–2.5 kbp of the promoter. Based on evidence from this figure elements upstream of 2.5 kbp seem to be inhibitory. Comparisons to the pGL3 empty luciferase vector were done using a Student’s t test. Data shown is the average of three biological replicates for at least three experiments, variation is shown as standard error of the mean (asterisk indicates mean greater than baseline, hash indicates mean less than baseline. *,#P ≤ 0.05, **,##P ≤ 0.01, ***,###P ≤ 0.001, ****,####P ≤ 0.0001)
