Fig. 3.
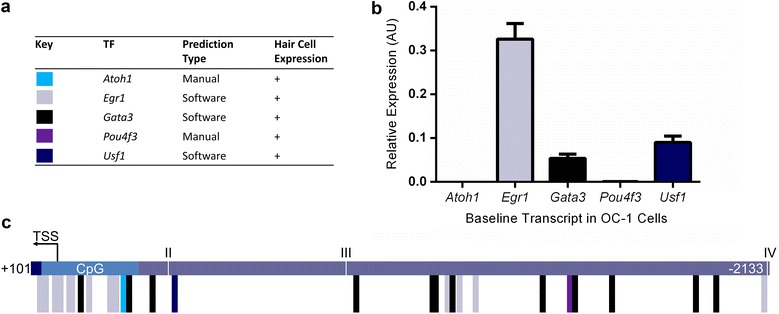
Transcription factors of interest and their binding sites in the αAtp2b2 [+101/−2133] promoter. Transcription factors were selected based on a multi-faceted in silico search. a Predicted transcription factor binding sites were found using software (TFBIND and MatInspector) or manually identified using published consensus sequences [17]. Hair cell expression was determined utilizing SHIELD data [27, 39]. SHIELD RNA-Seq data was collected from FACS sorted GFP expressing hair cells, transcripts with reads above zero were considered expressed and denoted in the table with a (+). b Transcription factor expression in OC-1 cells was determined utilizing qPCR and cross-referenced with published microarray data. EGR1, GATA3 and USF1 are expressed [25–27, 40]. Data is the average of three technical replicates for three biological replicates, variation is shown as standard error of the mean. c Predicted binding sites for ATOH1, EGR1, GATA3, POU4F3 and USF1 are shown. Note the clustering of predicted binding sites for transcription factors in the CpG island
