Fig. 4.
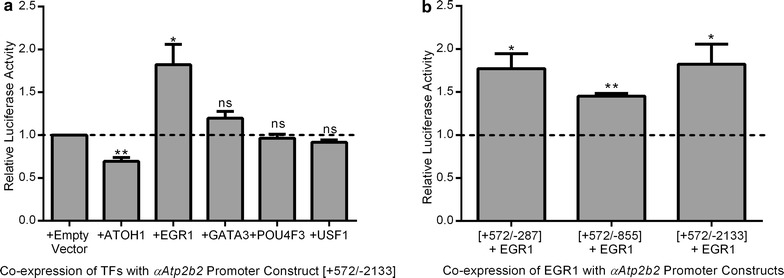
Co-expression of transcription factors with promoter constructs. a Luciferase activity of the αAtp2b2 promoter construct [+572/−2133] when co-expressed with transcription factor constructs or empty vector. Luciferase activity of the [+572/−2133] αAtp2b2 promoter construct co-expressed with transcription factor constructs was normalized to the luciferase activity of the [+572/−2133] promoter co-transfected with empty vector. Values were compared to a theoretical value of 1 utilizing a one-sample t test (*P ≤ 0.05, **P ≤ 0.01). b To narrow down the site of EGR1 activation in the αAtp2b2 promoter, three promoter truncations were assayed. The promoter truncation constructs were co-transfected with EGR1 or empty vector. Luciferase activity of promoter constructs co-transfected with EGR1 were normalized to luciferase activity of the promoter construct co-expressed with empty vector. Normalized values were compared to a theoretical value of 1 using a one sample t test (*P ≤ 0.05 and **P ≤ 0.01). Activation occurs over baseline promoter activity in all three constructs suggesting that the EGR1 binding site is contained in the region of the CpG island. Data for the αAtp2b2 promoter luciferase construct [+572/−2133] is the same in both a and b. Data shown is the average of three biological replicates for at least three experiments, variation is shown as standard error of the mean, dashed line represents the theoretical value of 1
