Abstract
Determinants of cellular tropism and receptor targeting lie within a short peptide in the Vr1 region of the envelope (Env) proteins of feline leukemia virus (FeLV) subgroups A and C. Libraries of FeLV Env proteins with random amino acid substitutions in the peptide were screened for their ability to deliver a marker gene to D17 and AH927 cells. Screening on D17 canine cells yielded D17-specific Env proteins that used the FeLV-C receptor. Screening on AH927 cells yielded Env proteins with a broader host range, with maximal titers on AH927 cells and similar or lower titers on other cells. These Env proteins used an unidentified non-FeLV receptor for entry. The A5 isolate obtained from the AH927 screen was readily concentrated to yield titers of 105 on human PC-3 prostate tumor cells. The sequence divergence observed among targeting peptides of library-selected Env proteins was greater than that found in parental FeLV isolates. Substitution analyses of a conserved R in the middle of the targeting peptide held constant during screening indicated that maximal titers were obtained only when R was present in both a D17 selected isolate and an AH927 selected isolate. The ability to isolate Env proteins with unique tropisms dependent on the cells on which the library is screened has direct implications for targeting gene delivery vectors.
The molecular mechanisms underlying the ability of divergent amino acid sequences within an Env protein to target a retrovirus to a particular receptor are not well understood. Receptor choice determinants for feline leukemia virus (FeLV) subgroups A and C (FeLV-A and FeLV-C) have been shown to reside in a short variable peptide sequence within Vr1 of the Env protein (17). Whereas FeLV-A preferentially infects feline cells, FeLV-C has a broader host range. By replacing a short peptide sequence within FeLV-A Env with the corresponding FeLV-C sequence, it was shown that both receptor usage and cellular tropism could be altered (17). FeLV-C uses a receptor that is homologous to organic anion transporters (14, 24). The receptor for FeLV-A has not yet been identified. Comparisons of Vr1 regions indicate that FeLV-C sequences are generally more similar to other FeLV-C sequences than FeLV-A sequences and vice versa (6, 16, 17, 22), suggesting that conserved portions of these variable sequences might determine receptor usage. Alternatively, these similarities could be due simply to natural evolutionary constraints that limit the possible diversity of sequence variation.
In order to bypass the evolutionary constraints and directly examine the extent of variability permitted within the FeLV-A and -C Env proteins, libraries of Env proteins containing random amino acid substitutions in the receptor-determining peptide were screened for the ability to deliver a gene to specific cells. The FeLV-A-derived Env protein used as the backbone for the library constructs was engineered to contain similarities to the FeLV-C Env within the receptor-targeting domain, including a three-amino-acid deletion and the conservation of the R of FeLV-C versus the K found within FeLV-A isolates. Selection of libraries randomized at 10 amino acid positions was previously successful in retargeting gene delivery (2, 3). Independent selection of the Env library has yielded isolates with unique properties. One variant specifically infected D17 canine osteosarcoma cells, using an FeLV-C receptor, while another isolate infected 143B human osteosarcoma cells, using a non-FeLV receptor (2, 3). The conclusions of the latter study with respect to retargeting gene therapy vectors were limited by the recovery of only a single retargeted Env protein that used a non-FeLV receptor.
Here the screening of the random FeLV Env library on D17 canine and AH927 feline cells is reported. This has allowed the isolation of multiple variants with identical target cell specificity and receptor usage. The peptides expressed on Env variants that infect D17 cells specifically via the FeLV-C receptor and others that preferentially infect feline and human cells through a non-FeLV receptor reveal little if any sequence conservation among them. However, mutagenesis of a central conserved R residue demonstrated the importance of this R in Env function. These results illustrate that the Vr1 peptide can undergo extensive variation and still target a given receptor. The isolation of two new variants that use a non-FeLV receptor extends the previous studies (2, 3), expanding the potential utility of the library-screening method for retargeting retroviral vectors.
MATERIALS AND METHODS
Cell lines and plasmids.
All fetal bovine serum was heat inactivated. D17 cells were maintained in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum. 293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 400 μg of G418 per ml. 293TCeB cells (2) expressing Moloney murine leukemia virus Gag and Pol proteins from the CeB plasmid (5), were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 400 μg of G418 and 6 μg of blasticidin S (ICN) per ml. TELCeB6 cells expressing a Ψ+ lacZ gene and Moloney murine leukemia virus Gag and Pol proteins (5) were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 6 μg of blasticidin S per ml. AH927 and 143B cells were maintained in Eagle's minimal essential medium with 10% fetal bovine serum. AH927CeB (2) and 143BCeB cells (3) were maintained in Eagle's minimal essential medium with 10% fetal bovine serum and 6 μg of blasticidin S per ml. PC-3 and DU-145 cells were maintained in RPMI 1640 with 25 mM HEPES, nonessential amino acids, and 10% fetal bovine serum. The plasmid library encoding Env proteins with random peptides inserted into the cell-targeting region has been described previously (3). These plasmids contain Ψ+ retroviral cassettes encoding the peptide-substituted env genes coupled to the neo gene encoding G418 resistance. pHIT-G was obtained from Michael Malim (7).
Library construction and screening.
Cell lines producing retroviral peptide libraries of two different complexities were constructed essentially as described (3). Briefly, 100 μg of each env library plasmid DNA (complexity = 3 × 107) and pHIT-G DNA or 300 μg of each were transiently transfected onto 10 10-cm dishes or 12 150-mm dishes of 293TCeB cells, respectively. The resulting supernatants containing retrovirus particles pseudotyped with the vesicular stomatitis virus G protein and packaging the library cassettes were filtered and used to inoculate 143BCeB cells in the presence of 8 μg of Polybrene per ml, which was removed the next day and replaced with fresh medium. After 2 days these cells were selected in 400 μg of G418 per ml, resulting in a population of cells constitutively producing retroviral particles. These particles expressed Env proteins with random peptide substitutions and packaged the env gene corresponding to the Env protein on the surface. The transfer of library cassettes from the transfected 293TCeB cells to the 143BCeB cells was done at a multiplicity of infection of <0.2 to ensure the transfer of only a single retrovirus per cell. Since two cassettes can be packaged by a single retrovirus, each resulting library producer cell could contain either one or two different peptide-substituted env genes. These libraries had complexities of 8 × 104 and 3 × 106 different peptides, depending on the amount of DNA that was transfected into the 293TCeB cells. The library of complexity 3 × 105 was described previously (3).
To screen the libraries, confluent producer cells were split 1:8 into drug-free medium, and 3 days later the supernatants were harvested and filtered through 0.45-μm filters before adding to target cells in the presence of 8 μM Polybrene. Target cells were AH927CeB or D17 cells. The CeB plasmid had been transfected into the AH927 cells to facilitate subsequent transfer of the env gene cassettes to other cell lines (2). After overnight incubation, the medium was changed to remove Polybrene, and 2 days later the cells were selected in the presence of 400 μg of G418 per ml. For further characterization, retroviral cassettes were transferred to a lacZ marker cell line by transfection of drug-resistant clones with pHIT-G (plus the Gag/Pol expression plasmid pCgp [8] for D17 clones), followed by transfer of the filtered supernatant to TELCeB6 lacZ cells in the presence of Polybrene 2 days later. Polybrene was removed the next day, and the cells were selected in 400 μg of G418 per ml 2 days later.
The screen of the highest complexity library on AH927CeB cells yielded 16 G418-resistant colonies; 15 of these were transferred to TELCeB6 cells for further screening. Of these, 11 gave titers of >103 per ml of the lacZ-carrying virus CFU on AH927 cells. Sequencing of targeting peptide-coding sequences from PCR-amplified genomic DNA from the TELCeB6 derivatives was done as previously described (2). Sequencing the targeting peptide coding regions of Env clones from the AH927 screen revealed that 10 isolates were identical. A5 is the prototype of these isolates and was examined in further detail. The single unique isolate was named A9a. Several clones identical to A5 were isolated from plates that had been separately inoculated with the library supernatant, indicating that they were the result of separate infectious events, confirming the reproducibility of the screen. The screen on D17 cells yielded 384 G418-resistant colonies from the low-complexity library and 561 from the high-complexity library. Only subsets of these colonies (47 and 45, respectively) were therefore transferred to the TELCeB6 marker cell line for further analyses. One Env derivative from the low-complexity screen and five from the high-complexity screen had titers of >103 per ml on D17 cells. Extrapolating these values back to the uncharacterized colonies and taking into account one cell division between library inoculation and drug selection, four variants were obtained from the low-complexity screen and 30 from the high-complexity screen.
Titering and interference assays.
To test the tropisms and titers of the selected Env proteins, confluent Env-expressing TELCeB6 derivatives were split 1:8 in the absence of drugs. Three days later supernatants were harvested, filtered, and used to inoculate cell lines on 2-mm gridded dishes at limiting dilutions in the presence of Polybrene. Titering results are the average ± standard deviation of two separate experiments, each carried out in duplicate. For the experiment shown in Table 2, L1 and A5 were concentrated by overnight centrifugation at 9,500 x g and 4°C; 115 ml of supernatant was reduced to 1 ml. For interference assays, target cells infected with FeLV-A, FeLV-B (90Z Env), FeLV-C, and 4070A were constructed as previously described (2, 3) as were control TELCeB6 cells expressing FeLV-A, FeLV-B (90Z), FeLV-C, and 4070A Env proteins. The interference assays reported in the tables are representative of two separate experiments carried out in duplicate.
TABLE 2.
Tissue specificity of A5 on human tumor cell lines
| Enva | Titer (lacZ CFU/ml) on target cells
|
||
|---|---|---|---|
| 143B | PC-3 | DU145 | |
| A5 | 4.1 ± 2.5 × 103 | 3.9 ± 0.7 × 105 | 2 ± 1 × 102 |
| L1 | 3.6 ± 1.0 × 105 | <50 | <50 |
| 4070A | 2.6 ± 1.7 × 105 | 4.6 ± 2.2 × 104 | 6.1 ± 3.0 × 104 |
L1 and A5 were concentrated by centrifugation as described in Materials and Methods.
Mutagenesis of the central arginine.
In order to express R mutants of B82 and A5, oligonucleotides encoding the region encompassing the targeting peptides were synthesized with all four nucleotides (represented by N) replaced in the codon coding for the central arginine residue: B82POS, 5′-GTGGGAGACACCTGGGAACCTACGGTATTCAGTGAGNNNCTCAGCGAATCCTCATCCTCCTCAAAATATGGA-3′; and A5POS, 5′-GTGGGAGACACCTGGGAACCCGCAGAGTAAGGGGCNNNGGTAATCAGTTAGGCTCCTCCTCAAAATATGGA-3′. These were then ligated into the BbsI-cut pRVL vector backbone after brief hybridization to the oligonucleotides RAN5F and RAN3F as previously described (2) for construction of the random library. The ligation mix was electroporated into Electromax DH10B competent cells (Gibco-BRL), and clones were picked for sequencing to confirm the ligation junctions and determine the amino acid substitution. Qiaquick minipreps (Qiagen) of the constructs were then transfected into 293TCeB cells with pHIT-G by using calcium chloride (Stratagene). This resulted in the production of retroviral particles expressing the vesicular stomatitis virus G protein on their surfaces and packaging the R mutant genes. After 0.45-μm filtration, the supernatants were used to infect TELCeB6 cells in the presence of 8 μg of Polybrene per ml overnight. Two days later cells were selected in G418 for cells that had taken up the R mutant genes. A5 env genes with W or D substitutions for the central R were constructed similarly with an oligonucleotide identical to A5POS except that the degenerate central position was replaced by TGG for W and GAT for D.
RESULTS
Peptide library screening.
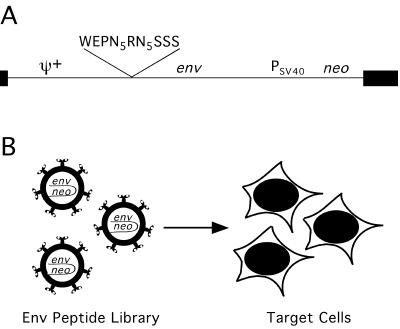
Libraries of FeLV-A Env derivatives containing random peptides in the receptor-determining region were expressed on the surfaces of retroviral particles as previously described (3). Each particle packages a cassette (Fig. 1A) encoding the Env protein expressed on its surface (Fig. 1B). If an Env protein containing an appropriate peptide is able to mediate gene delivery to a target cell, the target cell will become drug resistant (Fig. 1B). G418-resistant colonies resulting from such transfers can then be analyzed to further characterize the transferred env genes.
FIG. 1.
Peptide library screening strategy for targeting retroviruses. (A) The Ψ+ retroviral vector expressing the library of peptides in the receptor-targeting region of the Env protein is bicistronic. Black boxes are long terminal repeat (LTR) sequences. The sequence surrounding the 10 randomized amino acids denoted by N is shown. W is amino acid 52 of the mature 61E FeLV-A Env protein (6). The neo resistance gene is expressed from the simian virus 40 (SV40) promoter. (B) Retroviruses carrying the peptide library env gene cassettes and expressing corresponding peptide-substituted Env proteins on their surfaces were tested for cell targeting. Successful targeting leads to drug resistance gene transfer, followed by characterization of the linked env gene.
Libraries of three different complexities were screened on AH927CeB feline and D17 canine target cells. Env-expressing retroviral cassettes from resultant drug-resistant colonies were then transferred to TELCeB6 lacZ marker packaging cells in order to confirm and evaluate the functionality of selected variants. Variants with >103 TELCeB6 lacZ CFU per ml on the original target cells were characterized in further detail. Two such variants were isolated from the screen on AH927CeB cells with the highest-complexity library, but none was isolated from the library of intermediate complexity (Table 1). Hundreds of G418-resistant colonies were obtained from the screens of the lowest- and highest-complexity libraries on D17 cells. Therefore, only a subset of these colonies were characterized further. Of these, only one clone yielded >103 CFU per ml from the lowest-complexity screen, whereas five were obtained from the high-complexity screen. Extrapolating these numbers to the total yield of drug-resistant colonies suggests that 4 and 30 variants with >103 TELCeB6 lacZ CFU per ml were obtained from the lowest- and highest-complexity libraries, respectively (see Materials and Methods for details). Thus, more highly functional variants were obtained from higher-complexity libraries in both the AH927 and D17 screens.
TABLE 1.
Summary of Env libraries and target cells
| Library complexity | No. of functional variants obtaineda
|
|
|---|---|---|
| D17 | AH927 | |
| 8 × 104 | 4* | ND |
| 3 × 105 | ND | 0 |
| 3 × 106 | 30* | 2 |
Number of different Env variants obtained by screening libraries of various complexities on the indicated cell lines. Only variants with titers of >103 lacZ CFU per ml on the indicated cell lines are included. *, Values extrapolated from the subset of colonies tested. ND, not determined.
Targeted cell determines cellular tropism.
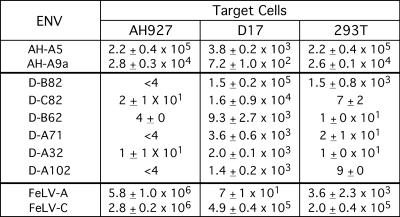
Isolates with titers >103 per ml on the original screened cell line were then tested further for their ability to infect AH927 feline, D17 canine, and 293T human cells (Fig. 2). The A5 and A9a isolates obtained by screening on AH927CeB cells had broad host ranges, infecting AH927 and 293T cells at similarly high levels (on the order of 104 to 105 per ml) and, to a lesser extent, D17 cells (102 to 103). All of the isolates obtained by screening on D17 cells maintained the highest titer on D17 cells. With one exception, B82, infection on AH927 or 293T cells was minimal for D17-selected isolates. B82 maintained a titer of 105 on D17 cells but also gave a lower titer of approximately 103 on 293T cells. The host ranges of the selected Env viruses are in contrast to the parental FeLV-A, which preferentially infects AH927 cells, and the related FeLV-C, which infects all three cell lines at similarly high levels. Thus, the tropisms of the library-derived Env proteins depended the cell line on which the library was screened.
FIG. 2.
Tropism of library-derived Env proteins is influenced by the cell line on which the library is screened. Library-derived Env proteins were expressed on retroviral particles produced from the TELCeB6 lacZ packaging cell line (5) and titered on feline AH927, canine D17, and human 293T cells. Env proteins from the library screen on AH927 cells are indicated by the AH prefix, those obtained from the screen on D17 cells are indicated by the D prefix. Results with wild-type FeLV-A and -C Env proteins are shown for comparison. Titers are TELCeB6 lacZ CFU per ml.
The host range and tissue specificity of the A5 Env was then analyzed on three additional human cell lines, PC-3 and DU145 prostate cells and 143B osteosarcoma cells (Table 2). PC-3 cells were derived from a bone metastasis (11), and DU145 cells were derived from a brain metastasis in a patient with prostate cancer (23). In agreement with earlier results demonstrating that a library-derived Env protein was stable to centrifugation (3), the A5 virus could be concentrated 100-fold by centrifugation, allowing titers on the order of 105 on PC-3 cells, 103 on 143B cells, and 102 on DU145 cells (Table 2). These titers were compared to the previously identified L1 isolate, which was selected on 143B cells and showed only limited infection on a broad range of cell lines (3), including PC-3 and DU145 cells. Thus, variants with different tropic properties on human cells were derived from two separate screens.
D17-targeted Env proteins use the FeLV-C receptor.
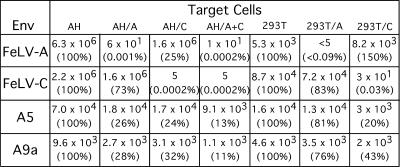
The receptor usage of the D17-targeted Env proteins was examined by receptor interference. Expression of an Env protein within a cell can block the subsequent infection of a virus utilizing the same viral receptor. Viral interference of the B82 and C82 derivatives, which gave the highest titer on D17 cells (Fig. 2), was determined. These two D17-specific Env proteins were tested for interference on FeLV-C-infected D17 cells (Table 3). A titer reduction of greater than 99% was considered significant interference (4, 15). The titers of the B82 and C82 isolates were reduced to 0.08% and 0.25% of their levels on uninfected D17 cells, respectively, when the cells were infected with FeLV-C. This indicates that they were using the FeLV-C receptor for viral entry. It was not possible to evaluate interference by FeLV-A due to the low titer of FeLV-A on this cell line. Similarly, it was not possible to reliably evaluate receptor usage by the other D17-specific Env proteins by the interference assay due to their lower titers and the high background (3 × 101) of non-Env-mediated gene transfer (Table 3).
TABLE 3.
Interference analyses of D17-screened Env proteins
| Env | Titer (lacZ CFU/ml) on target cellsa
|
|
|---|---|---|
| D17 | D17/FeLV-C | |
| FeLV-C | 1.5 × 106 | 3 × 101 (0.002) |
| B82 | 3.9 × 104 | 3 × 101 (0.08) |
| C82 | 2.0 × 104 | 5 × 101 (0.25) |
| None | 1 × 101 | 3 × 101 |
Values in parentheses are the titer on infected cells as a percentage of that on uninfected cells.
AH927-targeted Env proteins use a non-FeLV receptor.
The receptor usage of AH927-targeted A5 and A9a Env proteins was also examined by receptor interference assays. In this case, infection of AH927 and 293T cells by FeLV-A and FeLV-C did not prevent subsequent infection by either A5 or A9a (Fig. 3). AH927 cells doubly infected with FeLV-A and FeLV-C also remained sensitive to superinfection by A5 and A9a. These results indicate that A5 and A9a are not using either the FeLV-A or FeLV-C receptor. Similarly, neither FeLV-B nor amphotropic murine leukemia virus 4070A interfered with subsequent infection by A5 or A9a (Fig. 4A), indicating that A5 and A9a were not using the Pit1 or Pit2 receptor (1, 19, 25, 26). The nonreciprocal interference pattern observed between FeLV-B and 4070A on feline cells was due to the use of both Pit1 and Pit2 by FeLV-B (1, 19) and the exclusive use of Pit2 by 4070A (26). On human 293T cells, however, FeLV-B uses only Pit-1 (20, 25) and 4070A uses only Pit-2 (20, 26). Thus, nonreciprocal interference was not observed between FeLV-B and 4070A on 293T cells (Fig. 4A).
FIG. 3.
Library-derived A5 and A9a proteins do not use FeLV-A or FeLV-C receptors. Interference assays were used to examine receptor usage. lacZ transduction mediated by A5 or A9a on either AH927 (AH) or 293T cells infected with FeLV-A (A) and/or FeLV-C (C) was examined and compared with titers obtained on uninfected cells. The titer on infected cells as a percentage of that on uninfected is given in parentheses. Titers are TELCeB6 lacZ CFU per ml. Assays with 293T cells and AH927 cells were performed on different days and are each representative of two separate experiments.
FIG. 4.
(A) Library-derived A5 and A9a proteins do not use Pit-1 or Pit-2 receptors. Interference assays were performed on AH927 (AH) and 293T cells infected with either FeLV-B (B) or amphotropic murine leukemia virus 4070A (4070). AHCeB/A5 cells express the A5 Env protein. The titer on infected cells as a percentage of that on uninfected is given in parentheses. Titers are TELCeB6 lacZ CFU per ml. Assays with 293T cells and AH927 cells were performed on different days and are each representative of two separate experiments. (B) A5 Env expression does not interfere with FeLV-A or FeLV-C entry. Experimental details are as described for panel A.
In order to see whether A5 and A9a were using the same receptor, the original G418-resistant AH927CeB cells expressing A5 Env, obtained from the library screen, were used as target cells for interference assays. A5 expression in these cells prevented superinfection by both A5 and A9a, indicating that A5 and A9a were using the same receptor (Fig. 4A). FeLV-A, -B, and -C and 4070A were all able to infect A5 Env-expressing cells (Fig. 4A and B), confirming that these viruses were not using the same receptor as A5.
Targeting peptide sequences show no homology to each other.
In Fig. 5, the sequences of the targeting peptide regions of the two AH927-selected and six D17-selected Env proteins are compared with those from wild-type FeLV-A and -C Env proteins (6, 16, 17, 22). In the middle line of the figure, each randomized residue is indicated by an X and numbered 1 through 10. A5 and A9a were derived from the screen on AH927 cells. Clone C82 is the variant isolated from the low-complexity library screen on D17 cells. EF is a D17-specific isolate that was isolated from a previous screen on AH927 cells (2). The rest of the variants were isolated from the high-complexity screen on D17 cells. The library-derived sequences are listed in order of titer, with the highest-titer variants at the top of each group (AH927 and D17).
FIG. 5.
Library-derived targeting peptide sequences show little if any homology to each other or to wild-type FeLV-A and FeLV-C isolates. Targeting peptide sequences of wild-type isolates are shown at the top of the figure. Library-derived peptides are at the bottom of the figure, below the sequence of the peptide region randomized for library screening. Randomized residues are denoted by X and are numbered from 1 to 10. Dots indicate gaps in the sequence. AH927 indicates selection on AH927 cells. D17 indicates selection on D17 cells except for the previously reported EF isolate, which was selected on AH927 cells but specifically infects D17 cells (2).
There are no identities between the A5 and A9a targeting peptides at any position. In fact, the only conservative substitution is at position 6, where A5 has a G and A9a has an A. Otherwise, the residues at each position are quite different. Minimally, we know that A5 and A9a are within the same interference group, indicating functional similarity although no apparent sequence similarity.
For the D17-targeting peptides, it is the diversity of the sequences rather than the similarity that is striking. The most interesting homologies are the S/T substitutions at positions 1, 4, 9, and 10 in EF and B82, two isolates that maintain a titer of 105 on D17 cells. However, these are all replaced by other, quite different amino acids in both C82 and B62, which maintain a titer of 104 on D17 cells. Within the D17-targeted peptides, Ser is found two out of seven times at positions 4 and 6, Tyr twice at position 4, and Arg twice at position 1 and three times at position 7. However, overall the selected peptides are as dissimilar from each other as they are from the naturally occurring FeLV-A and FeLV-C isolates. The relative sequence conservation among the wild-type isolates is remarkable compared with the degeneracy of the library-derived sequences. While it is possible that all seven of the D17-targeted Env proteins use the FeLV-C receptor, this has only been directly demonstrated for the most highly functional variants (EF, B82, and C82). Limiting focus to these three proteins alone, it is clear that there is no specific motif or residue that might be responsible for targeting them to the canine FeLV-C receptor.
Arginine is optimal at the central position.
The conservation of a positively charged residue in the middle of the receptor-targeting domain of FeLV-A and FeLV-C and the retargeted Env proteins raised the issue of the significance of this residue for Env function. In order to address this question, the R was mutated in both the B82 and A5 Env proteins, and the effects of the change on titer were examined. In the case of the D17-targeted B82 (Table 4), several different amino acids were able to replace R. Although R was the optimal amino acid at this position, H, D, and N functioned as well as or better than the conservative K substitution. Hydrophilic amino acids (S, T, and Y) had slightly lower titers than the K substitution. Hydrophobic substitutions and P or C were barely functional, if at all.
TABLE 4.
Effects of changing the central R residue
| Substitution | Titera (lacZ CFU/ml)
|
|
|---|---|---|
| B82 substitution | A5 substitution | |
| R | 1.6 ± 0.2 × 105 | 1.6 ± 0.8 × 105 |
| H | 7.5 ± 0.2 × 104 | ND |
| D | 3.5 ± 0.4 × 104 | <8 |
| N | 9.3 ± 3.0 × 103 | <8 |
| K | 8.6 ± 3.4 × 103 | 6.5 ± 0.8 × 104 |
| Y | 5.05 ± 2.47 × 103 | ND |
| S | 2.8 ± 0.2 × 103 | ND |
| T | 1.8 ± 0.4 × 103 | <8 |
| W | 1.4 ± 0.3 × 102 | <8 |
| A | 1.2 ± 0.2 × 102 | <8 |
| L | 6.8 ± 2.8 × 101 | ND |
| V | 6.5 ± 2.5 × 101 | <8 |
| P | 6.4 ± 2.4 × 101 | ND |
| C | 4 ± 0 × 101 | ND |
| G | ND | <8 |
| I | ND | <8 |
| M | ND | <8 |
| Stop | 6 ± 2 | <8 |
B82 substitutions were tested on D17 cells, and A5 substitutions were tested on AH927 cells. ND, not determined.
In contrast, only the conservative change from R to K at the central position of A5 was allowed. D, N, A, V, T, G, I, M, and W were all detrimental (titers < 8 per ml) when tested on AH927 cells (Table 4). The K substitution had only a slightly lower titer (6.5 × 104 ± 0.8 × 104) than the R-containing A5 (1.6 × 105 ± 0.8 × 105). Similar results for A5 were observed on 293T cells, where only the K substitution was permitted (data not shown). Of particular interest is that titers were decreased on AH927 cells by over 4 orders of magnitude by D and N substitutions in A5, whereas the same substitutions in the D17-targeted B82 protein reduced B82 titers less than 20-fold. Thus, among other differences, replacement of a positive charge with a negative one was allowable in B82 but not in A5. The role that this residue plays in structure and function therefore appears different for the two Env proteins.
DISCUSSION
The results presented here demonstrate the ability of highly divergent peptide sequences to target FeLV Env proteins to the same receptor. The degree of divergence is far greater than the divergence observed in naturally occurring FeLV isolates. The similarity observed among natural isolates suggested that there might be important motifs that would determine receptor usage. Surprisingly, no motifs or conserved residues were identified after comparison of library-selected viruses that were within the same receptor interference groups. One possible explanation for the results reported here is that the highly divergent sequences obtained from a single round of library screening are not optimal for interaction with the target receptors. More-optimal sequences might show conservation such as occurs among wild-type isolates due to natural selective forces.
Control of receptor targeting by the hypervariable V3 loop of human immunodeficiency virus type 1 Env may provide an alternative analogy for the inability to identify receptor binding consensus sequences in the library-derived FeLV Env proteins. As with FeLV-A and -C Env and the library-derived variants, highly variable sequences within V3 determine whether human immunodeficiency virus type 1 uses one coreceptor or another for entry and the identification of consensus motifs has not been straightforward (9, 10, 21). Site-specific mutagenesis studies, however, have demonstrated the importance of several V3 residues in human immunodeficiency virus type 1 Env interactions with coreceptors (13, 18, 27). Whether or not these sequences interact directly with the coreceptor or influence coreceptor choice indirectly has not been conclusively determined (12), although direct interaction with conserved residues in V3 has been suggested (13, 18, 27). One of these residues is R298 at the base of the V3 loop, which is conserved among all human immunodeficiency virus type 1 and type 2 and simian immunodeficiency virus V3 sequences known to use CCR5 as a coreceptor (28). Replacement of this residue with non-positively charged amino acids eliminates human immunodeficiency virus type 1 infection (28). This is similar to the effect of substitution of the central conserved R in the AH927-selected A5 Env protein, although the precise role that these residues play in Env structure and function remains to be elucidated.
Quite interestingly, substitution analyses of both the D17-targeted B82 and the AH927-targeted A5 proteins indicated a strong dependence on the central conserved R of the targeting peptide for maximal entry. However, the range of substitutions allowed was quite distinct, indicative of different functional roles. Within the AH927 cell-targeting peptide of A5, only the R and the conservative K substitution were functional. In contrast, for the D17-targeted B82 Env, R is the optimal amino acid, but other charged or hydrophilic amino acids could replace it, yielding lower titers. Even a negatively charged amino acid could substitute for the positive charge. These data suggest that this residue may play an important role in hydrogen bond formation in B82 but might form a salt bridge in A5. Regardless of the mechanism, the central R of A5 is the only residue within the targeting region where there is a stringent requirement for conservation.
Although several different library-derived Env proteins targeted the FeLV-C receptor on D17 canine cells, no Env proteins targeted FeLV receptors on feline or human cells. Even a previously reported D17-specific Env protein (EF) isolated by selection on AH927 cells was only minimally functional on AH927 cells while it was highly functional with the FeLV-C receptor on D17 cells (2). A previous screen on human 143B cells also did not yield any FeLV receptor-targeted Env proteins (3). Since the overall goal of the library-screening approach is to target specific cell types for gene therapy, the backbone vector was designed as a structural hybrid between FeLV-A and FeLV-C Env proteins in order to minimize potential use of FeLV receptors. The length of the targeting peptide and the identity of the central, positively charged residue are those of FeLV-C Env rather than FeLV-A Env: 11 amino acids, including the central R of FeLV-C, versus 14 amino acids, including the central K residue of FeLV-A. However, the KY sequence four amino acids downstream of the peptide is more commonly found in FeLV-A Env. The mixed nature of the construct may therefore be incompatible with interaction with FeLV receptors expressed on the feline and human cells. This is consistent with earlier results examining hybrid FeLV-A/FeLV-C Env proteins with junctions within the Vr1 targeting peptide that were nonfunctional (17). It is possible that viral uptake on D17 cells does not require extensive direct contacts between this peptide domain and the canine receptor compared with the feline and human receptors. Residues outside the randomized peptide region may contact the receptor and play a greater role in binding of the FeLV-A Env backbone to D17 cells. This may be why it was possible to isolate several variants with FeLV receptors on these cells.
The inability to target human FeLV receptors by library screening is advantageous for the isolation of useful targeted Env proteins for gene therapy since otherwise targeted Env proteins would simply have the same tropisms as FeLV. Use of an Env backbone with a hybrid FeLV-A/C targeting region for library screening has been effective in the isolation of different Env proteins with distinct tropic properties on human cells with non-FeLV receptors. The previously reported L1 protein preferentially infects 143B and 293T cells (3). In contrast, A5 could be concentrated by centrifugation to a titer of over 105 on PC-3 cells, whereas L1-mediated gene delivery remained undetectable even after concentration on these cells. These different properties of A5 and L1 may be attributable to the fact that they were obtained by screening on different cell lines. The ability to isolate Env proteins with different human cell-targeting properties by using non-FeLV receptors is a promising result suggesting that other Env proteins with useful targeting properties may be obtained by screening additional Env libraries. Experiments to identify the receptors for A5 and L1 are in progress.
The properties of the isolated Env proteins are generally influenced by the cells on which the library is screened, although maximal titers are not always observed on the screening cell line (2, 3). At least three D17-specific variants that use the FeLV-C receptor and two variants (A5 and A9a) that use the same non-FeLV receptor on AH927 and 293T cells have been obtained. This suggests that certain cell surface proteins may be more readily targeted by Env library screening than others. Minor changes in the number and nature of randomized residues within the targeting peptide used for screening might be able to influence the receptor and cellular tropisms of library-derived variants. In order to target other receptors, it may be advantageous to bias codon usage against residues known to be important for interaction with the targeted receptors. For example, codons would be biased against R and K at the central position to target non-FeLV-C receptors on D17 cells or biased toward hydrophobic amino acids at the same position to circumvent the receptor used by A5 and A9a. Similar approaches could be applied to other positions to expand the range of possible target receptors.
The high complexity of the library compared with the small number of peptide sequences actually isolated indicates the stringent requirements for fully functional targeted peptides. These requirements are not limited solely to interactions with the receptor proteins but include other aspects of the selected targeting peptide's influence on Env structure and function. Every amino acid must be compatible with the folding pathway of the protein. It also must not interfere with other steps of the maturation of the protein, such as intracellular transport, incorporation into the viral particle, and proteolytic cleavage. Compatibility with receptor binding and virus-cell fusion are the final requirements for targeting peptide function. Thus, the combination of amino acids observed in selected peptides results from multiple selective forces. The lack of sequence conservation and the relative scarcity of functional targeting peptides highlight the complex nature of the intra- and intermolecular interactions driving the selection process. This type of information can be obtained most thoroughly and rapidly by screening peptide display libraries, where multiple amino acid substitutions can be made simultaneously.
Acknowledgments
This work is supported by awards 1ROI CA49932-11 from the National Institutes of Health and PC001347 from the Department of Defense to M.J.R.
We thank Taini Coriano, Bethany Barrow, and Carlos Rivera Adrovet for assistance in this project.
REFERENCES
- 1.Anderson, M. M., A. S. Lauring, S. Robertson, C. Dirks, and J. Overbaugh. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J. Virol. 75:10563-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bupp, K., and M. J. Roth. 2002. Altering retroviral tropism using a random-display envelope library. Mol. Ther. 5:329-335. [DOI] [PubMed] [Google Scholar]
- 3.Bupp, K., and M. J. Roth. 2003. Targeting a retroviral vector in the absence of a known cell-targeting ligand. Hum. Gene Ther. 14:1557-1564. [DOI] [PubMed] [Google Scholar]
- 4.Chesebro, B., and K. Wehrly. 1985. Different murine cell lines manifest unique patterns of interference to superinfection by murine leukemia viruses. Virology 141:119-129. [DOI] [PubMed] [Google Scholar]
- 5.Cosset, F.-L., Y. Takeuchi, J.-L. Battini, R. A. Weiss, and M. K. L. Collins. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donohue, P. R., E. A. Hoover, G. A. Beltz, N. Riedel, V. M. Hirsch, J. Overbaugh, and J. I. Mullins. 1988. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J. Virol. 62:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouchier, R. A. M., B. E. Meyer, J. H. M. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han, J.-Y., Y. Zhao, W. F. Anderson, and P. M. Cannon. 1998. Role of variable regions A and B in receptor binding domain of amphotropic murine leukemia virus envelope protein. J. Virol. 72:9101-9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman, T. L., and R. W. Doms. 1999. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 16:57-65. [DOI] [PubMed] [Google Scholar]
- 10.Jensen, M. A., F.-S. Li, A. B. van't Wout, D. C. Nickle, D. Shriner, H.-X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaighn, M. E., K. S. Narayan, Y. Ohnuki, J. F. Lechner, and L. W. Jones. 1979. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Investig. Urol. 17:16-23. [PubMed] [Google Scholar]
- 12.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaBrosse, B., C. Treboute, A. Brelot, and M. Alizon. 2001. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 75:5457-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quigley, J. G., C. C. Burns, M. M. Anderson, E. D. Lynch, K. M. Sabo, J. Overbaugh, and J. L. Abkowitz. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093-1099. [PubMed] [Google Scholar]
- 15.Rein, A. 1982. Interference grouping of murine leukemia viruses: A distinct receptor for the MCF-recombinant viruses in mouse cells. Virology 120:251-257. [DOI] [PubMed] [Google Scholar]
- 16.Riedel, N., E. A. Hoover, P. W. Gasper, M. O. Nicolson, and J. I. Mullins. 1986. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, FeLV-C Sarma. J. Virol. 60:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigby, M. A., J. L. Rojko, M. A. Stewart, G. J. Kociba, C. M. Cheney, L. J. Rezanka, L. E. Mathes, J. R. Hartke, O. Jarrett, and J. C. Neil. 1992. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J. Gen. Virol. 73:2839-2847. [DOI] [PubMed] [Google Scholar]
- 18.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 19.Rudra-Ganguly, N., A. Ghosh, and P. Roy-Burman. 1998. Retrovirus receptor PiT-1 of the Felis catus. Biochim. Biophys. Acta 1443:407-413. [DOI] [PubMed] [Google Scholar]
- 20.Sommerfelt, M. A., and R. A. Weiss. 1990. Receptor interference groups of 20 retroviruses plating on human cells. Virology 176:58-69. [DOI] [PubMed] [Google Scholar]
- 21.Speck, R. F., K. Wehrly, E. J. Platt, R. E. Atchison, I. F. Charo, D. Kabat, B. Chesebro, and M. A. Goldsmith. 1997. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J. Virol. 71:7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart, M. A., M. Warnock, A. Wheeler, N. Wilkie, J. I. Mullins, D. E. Onions, and J. C. Neil. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J. Virol. 58:825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stone, K. R., D. D. Mickey, H. Wunderli, G. H. Mickey, and D. F. Paulson. 1978. Isolation of a human prostate carcinoma cell line (DU 145). Int. J. Cancer 21:274-281. [DOI] [PubMed] [Google Scholar]
- 24.Tailor, C. S., B. J. Willett, and D. Kabat. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J. Virol. 73:6500-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi, Y., R. G. Vile, B. Simpson, B. O'Hara, M. K. L. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Zeijl, M., S. V. Johann, E. Closs, J. Cunningham, R. Eddy, T. B. Shows, and B. O'Hara. 1994. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 91:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, W.-K., T. Dudek, M. Essex, and T.-H. Lee. 1999. Hypervariable region 3 residues of HIV type 1 gp120 involved in coreceptor utilization: Therapeutic and prophylactic implications. Proc. Natl. Acad. Sci. USA 96:4558-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, W.-K., T. Dudek, Y.-J. Zhao, H. G. Brumblay, M. Essex, and T.-H. Lee. 1998. CCR5 coreceptor utilization involves a highly conserved arginine residue of HIV type 1 gp120. Proc. Natl. Acad. Sci. USA 95:5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]