Figure 5.
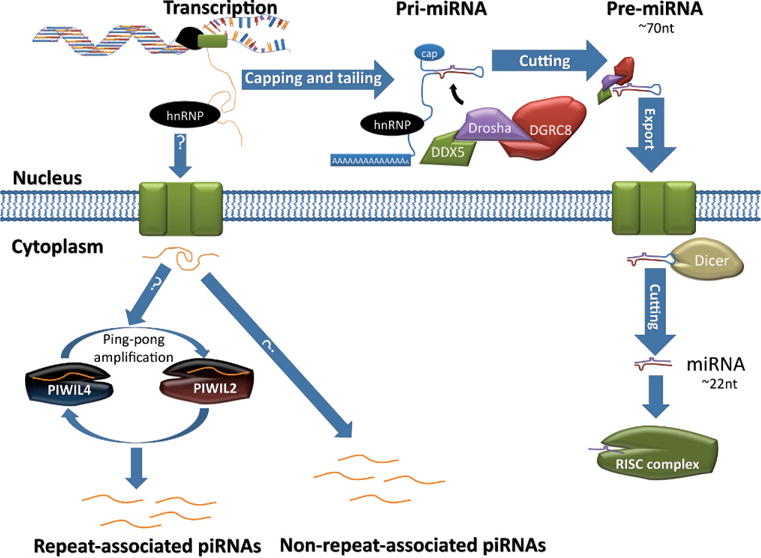
Biogenesis of microRNAs (miRNA) and PIWI-interacting RNAs (piRNA). DNA is transcribed into RNA by RNA polymerase II/III, and the resulting primary transcript (pri-miRNA) contains a hairpin structure that is recognized and cleaved by a microprocessor complex consisting of DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (DDX5), DGRC8, and DROSHA. This results in a ~70-nucleotide (nt) strand, called precursor miRNA (pre-miRNA), which retains its hairpin structure. Pre-miRNA is exported, and in the cytoplasm it is recognized and processed by an RNase III enzyme, DICER. This results in two ~22-nt mature miRNAs, which can then be incorporated into the RNA-induced silencing complex (RISC). The pathways forming piRNAs are not yet elucidated. But piRNAs are likely derived from long single-stranded RNAs transcribed in the nucleus. Once in the cytoplasm the precursor piRNAs are then processed into ~30-nt piRNAs. It is known that repeat-associated piRNAs are derived from transcripts of repetitive elements (eg, transposons), and PIWIL2 and PIWIL4 mediate the so-called “ping-pong” amplification loop to generate a large quantity of repeat-associated piRNAs, which can then suppress the expression of those “mother” repetitive elements through methylation of these loci. How non–repeat-associated piRNAs are produced remains unknown.
