Table S1.
Pathway level analysis of circadian changes with HCC
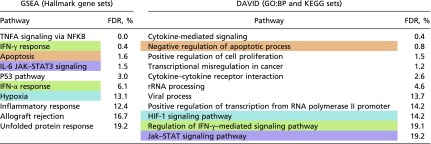 |
After temporal reconstruction with CYCLOPS, transcripts having lost rhythmicity in HCC were identified. This required (i) significant cycling in NC samples by the following criteria: (ii) the lack of significant cycling in HCC samples; (iii) the significance of tumor specific circadian parameters in a nested model; and (iv) a more than twofold reduction in amplitude. KEGG, Reactome, and GO biological (direct) processes sets that were overrepresented among those genes that lost rhythmicity in HCC are shown. Separately, genes were ranked by the change in amplitude when comparing HCC and NC samples. GSEA was used to identify pathways enriched with amplitude reduction. For both analyses, the false-discovery rate (FDR) as output from the corresponding programs are shown for each gene set. Gene sets highlighted in the same color represent overlapping physiological pathways.
