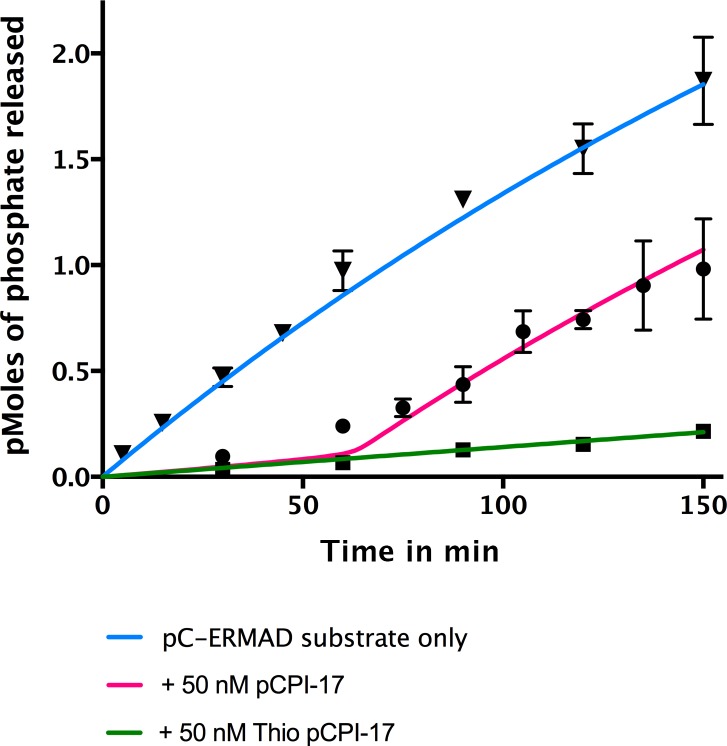
Figure 5. In vitro demonstration of the unfair competition mechanism for MLCP and pCPI-17.
Enzyme assays containing 0.25 nM MLCP and 1.4 μM C-ERMAD were performed at 30°C for the indicated times, either in the absence of CPI-17 (inverted triangles) or in the presence of 50 nM pCPI-17 (circles) or 50 nM thio(p)CPI-17 (squares). N = 3 for all data points; error bars represent standard errors of the mean. The data show that MLCP must first dephosphorylate pCPI-17 before it can target the C-ERMAD substrate. The blue and green lines are nonlinear regressions to the data in the absence of pCPI-17. The red line is the theoretical prediction based on the previously measured pCPI-17/MLCP kinetic constants as described in Supplementary file 1.