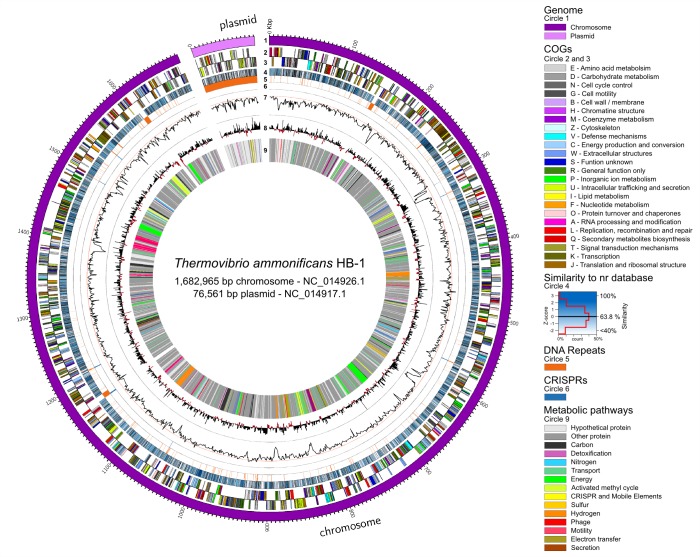
Figure 2. Thermovibrio ammonificans HB-1 genomic map highlighting main genomic features.
The circular chromosome and plasmid are drawn together. Several other repeats were identified in the chromosome (circle 5). From the outer circle: 1 – Dimension of the genome in base pairs (chromosome in violet, plasmid in pink); 2 and 3 – sense and antisense coding sequences colored according to their COGs classification (see legend in the figure); 4 – similarity of each coding sequence with sequences in the non-redundant database; 5 – position of tandem repeat in the genome. A total of 46 tRNA were identified, comprising all of the basic 20 amino acid plus selenocysteine; 6 – position of CRISPRs; 7 – GC mol% content, the red line is the GC mean of the genome (52.1 mol%); 8 – GC skew calculated as G+C/A+T; 9 – localization of coding genes arbitrarily colored according to the metabolic pathways reconstructed (see Figure 1). Colors are consistent trough the entire paper. Total dimension and accession number for the chromosome and plasmid are given.