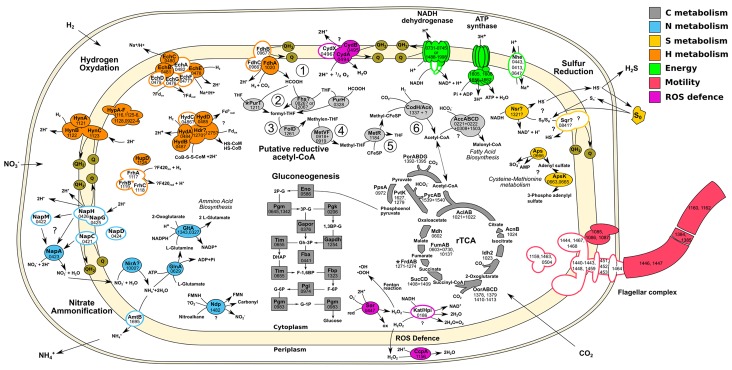
Figure 3. Central metabolism of T. ammonificans HB-1.
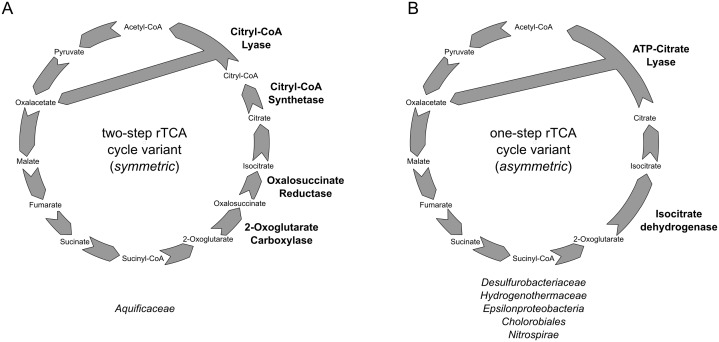
Enzyme names are reported together with the gene locus number (Theam_number). Primary compounds involved in reactions were also reported, however visualized reactions are not complete. Pathways were arbitrarily color coded according to their reconstructed function and are consistent throughout the paper. Solid shapes represent genes for which the enzyme was found in the proteome, while outlined shapes were only identified in the genome. Circled numbers 1 to 6 represent reaction numbers of the putative reductive acetyl-CoA pathway as described in the text. Abbreviations: NITRATE AMMONIFICATION: NapCMADGH – Periplasmic nitrate reductase complex; NirA – Putative nitrite reductase; AmtB – ammonia transporter; GlnA – L-Glutamine synthetase; GltA – Glutamate synthetase. HYDROGEN OXIDATION: HynABC – Ni-Fe Membrane bound hydrogenase; FrhACB – Cytosolic Ni-Fe hydrogenase/putative coenzyme F420 hydrogenase; HupD – Cyrosolic Ni-Fe hydrogenase maturation protease; HypA-F – Hydrogenases expression/synthesis accessory proteins; EchABCEF – Ech membrane bound hydrogenase complex; HydAB – Cytosolic Ni-Fe hydrogenases potentially involved in ferredoxin reduction; Hdr? – Missing heterodisulfide reductase CoB-CoM; FdhABC – Formate dehydrogenase. ENERGY PRODUCTION: NADH dehydrogenase and ATP synthetase are reported without the names of the single units; Nhe – Sodium/hydrogen symporter. SULFUR REDUCTION: Sqr – Putative sulfate quinone reductase involved in sulfur respiration; Nsr? – FAD/NAD nucleotide-disulphide oxidoreductase; Aps – Sulfate adenylyl transferase and kinase involved in assimilation of sulfur. FLAGELLAR COMPLEX: for simplicity single unit names are not reported. REDUCTIVE ACETYL-CoA: Fhs – Proposed reverse formyl-THF deformylase; PurHT – Formyltransferases (phosphoribosylaminoimidazolecarboxamide and phosphoribosylglycinamide) FolD – Methenyl-THF cyclohydrolase and dehydrogenase; MetVF – Methylene-THF reductase; MetR – Putative methyl-transferase; CodH – Carbon monoxyde dehydrogenase; AcsA – Acetyl-CoA ligase/synthase; AccABCD – Acetyl-CoA carboxylase. REDUCTIVE CITRIC ACID CYCLE: AclAB – ATP-citrate lyase; Mdh – Malate dehydrogenase; FumAB – Fumarate hydratase; FrdAB – Fumarate reductase; SucCD – Succinyl-CoA synthetase; OorABCD – 2-Oxoglutarate synthase; Idh2 – Isocitrate dehydrogenase/2-oxoglutarate carboxylase; AcnB – Aconitate hydratase; PorABDG – Pyruvate synthase; PycAB - Pyruvate carboxylase; PpsA – Phosphoenolpyruvate synthase water dikinase; PyK – Pyruvate:water dikinase. GLUCONEOGENESIS: Eno – Enolase; Pgm – Phosphoglycerate mutase; Pgk – Phosphoglycerate kinase; Gapor – Glyceraldehyde-3-phosphate dehydrogenase; Gapdh – Glyceraldehyde 3-phosphate dehydrogenase; Fba – Predicted fructose-bisphosphate aldolase; Tim – Triosephosphate isomerase; Fbp – Fructose-1,6-bisphosphatase I; Pgi – Phosphoglucose isomerase; Pgm – Phosphoglucomutase.
DOI: http://dx.doi.org/10.7554/eLife.18990.007