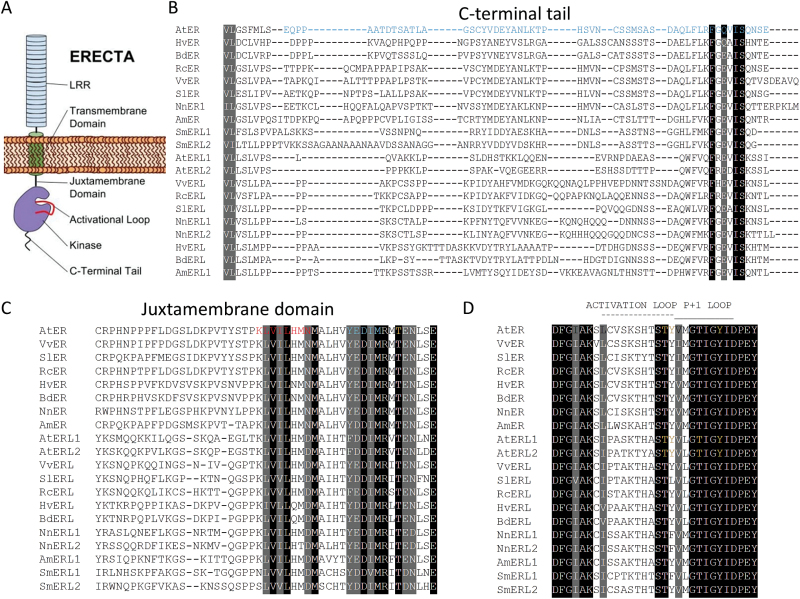
Fig. 1.
Comparison of amino acid sequences of ERECTA family proteins in different species. (A) Domain structure of the ERECTA receptor. (B–D) MAFFT alignment of the predicted amino acid sequences of ERECTA family genes from Arabidopsis thaliana (At), Vitis vinifera (Vv), Solanum lycopersicum (Sl), Ricinus communis (Rc), Hordeum vulgare (Hv), Brachypodium distachyon (Bd), Nelumbo nucifera (Nn), Amborella trichopoda (Am), and Selaginella moellendorffii (Sm). Residues that are identical among the sequences are shown with a black background, and those that are similar among the sequences are shown with a gray background. (B) The C-terminus. The blue residues have been deleted in pPZK110 and in pPZK111. (C) The juxtamembrane domain. The red residues have been deleted in pPZK104, the blue residues in pPZK105. Threonine in yellow has been substituted with Ala in pPZK102. (D) The activation loop. The predicted phosphorylation sites according to the Arabidopsis Protein Phosphorylation Site Database (PhosPht) are in yellow.