Highlight
Sequential action of kiwifruit CEN, BFT, and FT genes regulates growth, dormancy, and flowering. Two differentially expressed FT genes have different impacts on flowering and plant morphology.
Keywords: Actinidia chinensis, BFT, CEN, development, dormancy, flowering, FT, kiwifruit.
Abstract
Kiwifruit is a woody perennial horticultural crop, characterized by excessive vegetative vigor, prolonged juvenility, and low productivity. To understand the molecular factors controlling flowering and winter dormancy, here we identify and characterize the kiwifruit PEBP (phosphatidylethanolamine-binding protein) gene family. Five CEN-like and three BFT-like genes are differentially expressed and act as functionally conserved floral repressors, while two MFT-like genes have no impact on flowering time. FT-like genes are differentially expressed, with AcFT1 confined to shoot tip and AcFT2 to mature leaves. Both act as potent activators of flowering, but expression of AcFT2 in Arabidopsis resulted in a greater impact on plant morphology than that of AcFT1. Constitutive expression of either construct in kiwifruit promoted in vitro flowering, but AcFT2 displayed a greater flowering activation efficiency than AcFT1, leading to immediate floral transition and restriction of leaf development. Both leaf and flower differentiation were observed in AcFT1 kiwifruit lines. Sequential activation of specific PEBP genes in axillary shoot buds during growth and dormancy cycles indicated specific roles in regulation of kiwifruit vegetative and reproductive phenologies. AcCEN and AcCEN4 marked active growth, AcBFT2 was associated with suppression of latent bud growth during winter, and only AcFT was activated after cold accumulation and dormancy release.
Introduction
The phosphatidylethanolamine-binding proteins (PEBPs), initially identified in animals as Raf-1 kinase inhibitors (Yeung et al., 1999), provide a potential to fine-tune developmental regulatory mechanisms and present opportunities for improvement of agriculturally important traits (Jung and Müller, 2009; Zhang et al., 2010; Yeoh et al., 2011; Iwata et al., 2012; Putterill et al., 2013; Koskela et al., 2016). In angiosperms, this family consists of three phylogenetically distinct groups: the FLOWERING LOCUS T (FT)-like proteins (Kardailsky et al., 1999; Kobayashi et al., 1999), the TERMINAL FLOWER1 (TFL1)-like and CENTRORADIALIS (CEN)-like proteins (Bradley et al., 1996, 1997; Ohshima et al., 1997), and the MOTHER OF FT AND TFL1 (MFT)-like proteins (Mimida et al., 2001; Yoo et al., 2004). In addition, the BROTHER OF FT AND TFL1 (BFT)-like proteins (Yoo et al., 2010) represent a separate subclade within the CEN group.
FT- and TFL1/CEN-like genes have conserved roles in regulation of flowering, and the ancestral MFT-like genes (Hedman et al., 2009; Karlgren et al., 2011) have been associated with seed germination (Xi et al., 2010; Footitt et al., 2011; Nakamura et al., 2011). FT-like proteins can act as florigen (Chailakhyan, 1968; Zeevaart, 2008), moving in the phloem from leaves to the shoot apex in a highly regulated manner (reviewed in Putterill and Varkonyi-Gasic, 2016) to promote flowering. In contrast, TFL1/CEN-like proteins repress flowering and maintain the inflorescence meristem (Shannon and Meeks-Wagner, 1991; Ratcliffe et al., 1998, 1999; Conti and Bradley, 2007). This antagonistic activity (Shalit et al., 2009; Lifschitz et al., 2014) involves interaction with a common partner FD (Pnueli et al., 2001; Abe et al., 2005; Wigge et al., 2005; Danilevskaya et al., 2010; Hanano and Goto, 2011; Randoux et al., 2014) and is conferred by a divergent external loop and specific amino acids in positions critical for FT function (Hanzawa et al., 2005; Ahn et al., 2006). Other changes resulting in altered surface charge can affect function (Ho and Weigel, 2014), and some FT-like proteins act as repressors of flowering (Pin et al., 2010; Harig et al., 2012; Cao et al., 2016). In many plant species, numerous FT-like genes arose from gene duplication events (Pin and Nilsson, 2012), but only one or two act in flowering control, and FT-like roles besides flowering, such as vegetative growth and storage organ differentiation, have been reported (Hsu et al., 2011; Navarro et al., 2011; Lee et al., 2013), confirming a key role for PEBPs in co-ordination of general growth and development.
Many aspects of PEBP biology remain less well understood in woody perennial plants (Putterill and Varkonyi-Gasic, 2016). Recent studies of the most advanced woody perennial model poplar (Populus spp.) revealed conservation between molecular regulation of flowering in annual plants and seasonal phenology in trees (Ding and Nilsson, 2016). In addition to regulation of flowering, poplar FT- and TFL1/CEN-like genes are implicated in the regulation of vegetative to reproductive transition, growth and dormancy cycles, and the co-existence of vegetative and floral meristems on the same shoot (Böhlenius et al., 2006; Hsu et al., 2006, 2011; Rinne et al., 2011). Similarly, TFL1 is a key regulator of perennial growth and seasonality of flowering in strawberry and rose (Iwata et al., 2012; Koskela et al., 2012, 2016; Randoux et al., 2014; Rantanen et al., 2015), down-regulation of TFL1-like genes accelerated flowering in apple and pear (Kotoda et al., 2006; Freiman et al., 2012; Yamagishi et al., 2016), ectopic overexpression of a CEN homolog suppressed flowering in kiwifruit (Varkonyi-Gasic et al., 2013), and a connection of TFL1 accumulation with biennial bearing in apple has been established (Haberman et al., 2016).
On the other hand, constitutive or inducible expression of Arabidopsis or poplar FT genes has been attempted and used with some success to accelerate breeding (Srinivasan et al., 2012; Wenzel et al., 2013), although many questions regarding the FT gene, promoter, and feasibility of the approach remain (Zhang et al., 2010). Remarkably, only a handful of endogenous FT genes have been functionally characterized by transgenic approaches in woody perennial species; expression under a strong constitutive promoter resulted in extremely early flowering in trifoliate orange, poplar, apple, and blueberry (Endo et al., 2005; Böhlenius et al., 2006; Hsu et al., 2006; Kotoda et al., 2010; Yamagishi et al., 2011, 2014; Song et al., 2013), and only one example of graft-transmissible early flowering has been reported in a small woody shrub, Barbados nut (Jatropha) (Ye et al., 2014). Even less is known about functional diversification of FT genes in woody perennials. Ectopic expression implied distinctive roles in reproductive and vegetative growth for poplar FT paralogs (Hsu et al., 2011), not yet confirmed by paralog-specific silencing; expression of only one of two FT genes in apple and one of three FT genes in citrus has been associated with transition to flowering (Nishikawa et al., 2007; Kotoda et al., 2010), and expression of a single grape FT gene could be detected irrespective of flowering (Sreekantan and Thomas, 2006; Carmona et al., 2007). Furthermore, some FT genes failed to induce early flowering in woody perennials, despite their ability to promote flowering in model annuals, while affecting dormancy and leaf senescence in apple (Freiman et al., 2015) or shoot vigor in kiwifruit (Varkonyi-Gasic et al., 2013).
Although the ability to regenerate transgenic woody perennials efficiently remains an obstacle, development of new technologies to induce gene-specific biallelic homozygous mutations in trees (Fan et al., 2015) and specifically manipulate members of the PEBP family holds great promise in accelerating the breeding process and improving the architecture and productivity of woody perennials of horticultural importance. Detailed functional characterization of PEBP genes is the necessary first step to improve selection of gene candidates likely to have a key role in maturation and flowering.
Kiwifruit (Actinidia) is one of the most recently domesticated horticultural crops, with a short history of breeding (Datson and Ferguson, 2011), limited genetic resources, and little understanding of molecular regulation or the genetic diversity in flowering responses to internal and environmental signals. Commercial kiwifruit cultivars belong to closely related A. chinensis and A. deliciosa, which, together with other members of the Actinidia genus, are woody perennial vines from the order Ericales in the asterid lineage, with features of development specific to temperate woody perennials, including prolonged juvenility, growth spread over two seasons (Brundell, 1975a, b), and a period of winter chilling required to resume growth after winter dormancy (Brundell, 1976). Excessive vegetative growth and a small number of flowers, resulting in low fruit yield and high orchard costs, are key targets in kiwifruit improvement strategies. Flower differentiation occurs during budbreak in spring, from pre-established but undifferentiated overwintering primordia (Walton et al., 1997, 2001; Varkonyi-Gasic et al., 2011). Inflorescences develop in the lower leaf axils of the shoots that emerge after sufficient winter chilling, and photoperiod has no clear role in regulation of flowering in spring (Snelgar et al., 2007).
An FT and a CEN gene have been described in kiwifruit previously (Varkonyi-Gasic et al., 2013), but AcFT failed to induce precocious flowering in transgenic kiwifruit and it remained unclear if other PEBP genes might be involved in regulation of kiwifruit maturity, architecture, and phenology. The completion of the draft genome sequence of A. chinensis (Huang et al., 2013) provided the opportunity to identify additional sequences belonging to the kiwifruit PEBP family. In this study, we identified 13 Actinidia PEBP genes, five belonging to the TFL1/CEN clade, three each belonging to the BFT and FT clade, and two belonging to the MFT clade. All genes with the exception of the previously characterized AcFT and AcCEN (Varkonyi-Gasic et al., 2013) were characterized here using a combination of expression analysis and functional studies in transgenic Arabidopsis. Two of the previously undescribed FT-like genes were further characterized in more detail and their role in activation of flowering was evaluated in transgenic kiwifruit. The information provided help to better understand the diversity and biological function of the PEBP family in woody perennial plants and identifies candidate targets for improvement of kiwifruit architecture, flowering time, and plant determinacy.
Materials and methods
Plant materials
Kiwifruit plant material was sampled from female kiwifruit cultivars, ‘Hort16A’ (Actinidia chinensis Planch., recently classified as A. chinensis Planch. var. chinensis) and ‘Hayward’ [A. deliciosa (A. Chev.) C.F. Liang et A.R. Ferguson, recently classified as A. chinensis var. deliciosa (A. Chev.) A. Chev.]. Sampling for gene expression analysis was described previously (Wu et al., 2012; Varkonyi-Gasic et al., 2013). Briefly, root, stem, leaf, flower, and fruit tissue were collected from mature plants grown in the Plant & Food Research orchard near Kerikeri, New Zealand in the spring and summer season of 2005–2006, basal and distal leaves from mature ‘Hort16A’ plants grown in the Plant & Food Research orchard near Te Puke in spring of 2010, and axillary bud samples from field-grown ‘Hayward’ plants grown in a commercial orchard near Hamilton, New Zealand during the 1995–1996 season. Amplification of full-length coding sequences was performed on a set of ‘Hort16A’ cDNA described in Ledger et al. (2010). Additional leaf samples (L1–L8) were collected from 1-year-old ‘Hort16A’ plants grown in the Plant & Food Research glasshouse in Auckland, New Zealand in spring 2013. Axillary bud samples used in RNA-seq experiments were collected at monthly intervals from A. chinensis plants grown in the Plant & Food Research orchard near Te Puke, New Zealand in the 2014–2015 season. Terminal buds were sampled from the same plants in January and March 2015. Three plants (biological replicates) were used and ~10 buds were sampled at each time point, with the exception of the March terminal bud sample, where only two plants were available. Sampling of tissues was performed at mid-day to avoid variation.
Isolation of the genes and vector construction
Gene-specific oligonucleotide primers were designed based on available sequence data (Huang et al., 2013) (see Supplementary Table 1 at JXB online). Full-length coding sequences were amplified from A. chinensis ‘Hort16A’ cDNA, cloned into the pUC19 vector, and verified by sequence analysis (GenBank accession nos KX611594–KX611604). After addition of attB sites by PCR, each gene was recombined using Gateway into pDONR221, verified by sequence analysis, then recombined into pHEX2 (Hellens et al., 2005), placing each cDNA between the Cauliflower mosaic virus (CaMV) 35S promoter and the ocs 3' transcriptional terminator. The modified AcFT1 and AcFT2 sequences with added attB sites were synthesized by GenScript (http://www.genscript.com) and recombined into pHEX2 as above. For SUC2:AcFT1 and SUC2:AcFT2 constructs, the cDNAs were re-amplified to include appropriate restriction sites and cloned as SpeI–XbaI fragments into pSAK778-SUC2 to replace AcFT (Varkonyi-Gasic et al., 2013). The 35S:GUS vector control was constructed previously (Drummond et al., 2009). All resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. 35S:AcFT1 and 35S:AcFT2 were also transformed into A. tumefaciens strain EHA105 by electroporation.
Phylogeny
Predicted protein sequence alignment and phylogenetic analyses were conducted using Vector NTI (Invitrogen) CLUSTAL W, and phylogeny was constructed using a minimum evolution phylogeny test and 1000 bootstrap replicates in MEGA version 5.2 (Tamura et al., 2011).
Plant transformation and growth
Agrobacterium tumefaciens-mediated Arabidopsis transformation was performed as described (Clough and Bent, 1998; Martinez-Trujillo et al., 2004), using constructs in A. tumefaciens strain GV3101. Seeds of transgenic plants were selected on half-strength Murashige and Skoog (1/2 MS) medium supplemented with kanamycin and placed in a growth room under a long-day (LD, 21 °C, 16/8 h light/dark) or short-day (SD, 21 °C, 8/16 h light/dark) regime. Plants were grown in soil using a standard potting mix. Transformation of A. chinensis ‘Hort16A’ was as previously described (Wang et al., 2007; Voogd et al., 2015), using A. tumefaciens strain EHA105.
RNA extraction and expression studies
Total RNA from Arabidopsis was isolated using the Trizol reagent (Invitrogen) and from kiwifruit using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA). Reverse transcription was performed using the QuantiTect Reverse Transcription Kit (Qiagen). Quantifications using real-time PCR were performed with the FastStart DNA Master SYBR Green I mix (Roche Diagnostics, Mannheim, Germany) using the LightCycler 1.5 instrument and the LightCycler Software version 4 (Roche Diagnostics). Oligonucleotide primers (Supplementary Table S1) were designed to produce amplification products of 100–150 nucleotides. The specificity of primer pairs was confirmed by melting curve analysis of PCR products and agarose gel electrophoresis followed by sequence analysis. The expression was normalized to Actinidia ACTIN (GenBank accession no. FG403300) or Arabidopsis ACT2 (At3g18780).
The axillary bud sequencing libraries were constructed according to the TruSeq RNA sample preparation guide (Illumina, San Diego, CA, USA) and subsequently sequenced by a HiSeq 2500 Sequencing System (Illumina) at Australia Genomics Resource Facility (Brisbane, Australia), obtaining paired-reads of 125 bp. Raw sequence data in FASTQ format were filtered to remove the adaptors, low-quality reads, and reads containing ambiguous nucleotides, resulting in an average of 13.8 million reads per library. As the PEBP genes are mostly misannotated in the draft kiwifruit genome and the genome is generally poorly annotated, the mapping was performed on the 13 sequences representing the kiwifruit PEBP gene family, using STAR version 2.5.2a (Dobin et al., 2013), and only uniquely mapped reads with no mismatches per paired alignment were used for gene expression studies. The gene expression unit was calculated as fragments per kilobase of transcript per million reads (FPKM).
Yeast two-hybrid assay
AcFT1 and AcFT2 coding regions were recombined from entry clones into the GATEWAY destination vectors pDEST32 (pBDGAL4, bait) and pDEST22 (pADGAL4, prey) (Invitrogen). The remaining clones were as described before (Varkonyi-Gasic et al., 2013). These vectors were individually introduced into Saccharomyces cerevisiae strains PJ69-4α (bait) and PJ69-4a (prey) (James et al., 1996) for selection on minimal medium plates (WO) lacking leucine (bait) or trytophan (prey), followed by mating on YPAD plates and double sequential selection on WO medium lacking both leucine and trytophan. Final screening was performed on medium lacking trytophan, leucine, and histidine and supplemented with 0, 1, 2, 5, or 25 mM 3-amino-1,2,4-triazole. Plates were incubated for 4 d at 20 °C, photographed, and scored for growth. Reciprocal tests were performed in duplicate for all combinations.
Results
The Actinidia PEBP gene family
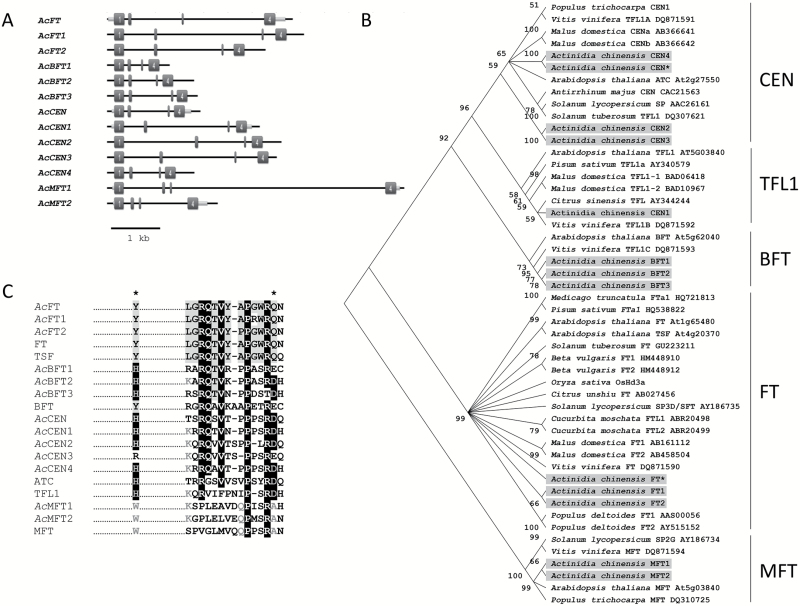
Searches for putative FT-, TFL1/CEN-, and MFT-like genes in the A. chinensis draft genome (Huang et al., 2013), combined with analyses of the Actinidia EST database (Crowhurst et al., 2008), revealed a total of 13 sequences representing the kiwifruit PEBP gene family (Fig. 1A). Five sequences with homology to TFL1/CEN, three with homology to related BFT, three with homology to FT, and two MFT-like sequences were identified, including previously characterized Actinidia FT (AcFT) and CEN (AcCEN) sequences (Varkonyi-Gasic et al., 2013). Amplification and sequencing were further deployed to confirm full-length coding sequences. Predicted protein sequences were used for phylogenetic analysis (Fig. 1B) and for analysis of amino acid positions critical for function. Like the previously described AcFT, newly identified kiwifruit FT-like proteins, annotated AcFT1 and AcFT2, have the conserved glutamine within the external loop (14 amino acids in segment B) and the adjacent tyrosine residue, critical for FT function (Hanzawa et al., 2005; Ahn et al., 2006), but divergence in the external loop of AcFT1 (G137R) was identified (Fig. 1C). In addition, a change of isoleucine at the position highly conserved across the PEBP proteins (I117F) and cysteine at a position mostly conserved across FT-like proteins (C164G) was identified in AcFT1 (Supplementary Fig. S1). Additional divergent amino acids between AcFT1 and AcFT2 were at positions not highly conserved across FT proteins. With the exception of AcCEN3 and AcBFT1, all kiwifruit CEN- and BFT-like proteins have the conserved residues equivalent to Arabidopsis His88 and Asp144, while in the equivalent positions both MFT-like sequences have tryptophan and alanine residues (Fig. 1C).
Fig. 1.
The Actinidia PEBP gene family. (A) Genomic organization of the genes. Dark gray boxes represent exons and light gray boxes represent untranslated region sequences (where available, based on RNA database searches). (B) Phylogram of plant PEBP proteins. The kiwifruit proteins are highlighted and previously described AcFT and AcCEN are indicated with an asterisk. (C) Partial amino acid alignment of Actinidia (Ac) and Arabidopsis PEBP proteins including the conserved segment B region.. Asterisks indicate Tyr85 (Y)/His88 (H) and Gln140 (Q)/Asp144 (D) residues distinguishing FT-like and TFL1-like members.
Differential expression of the Actinidia TFL1/CEN- and BFT-like genes
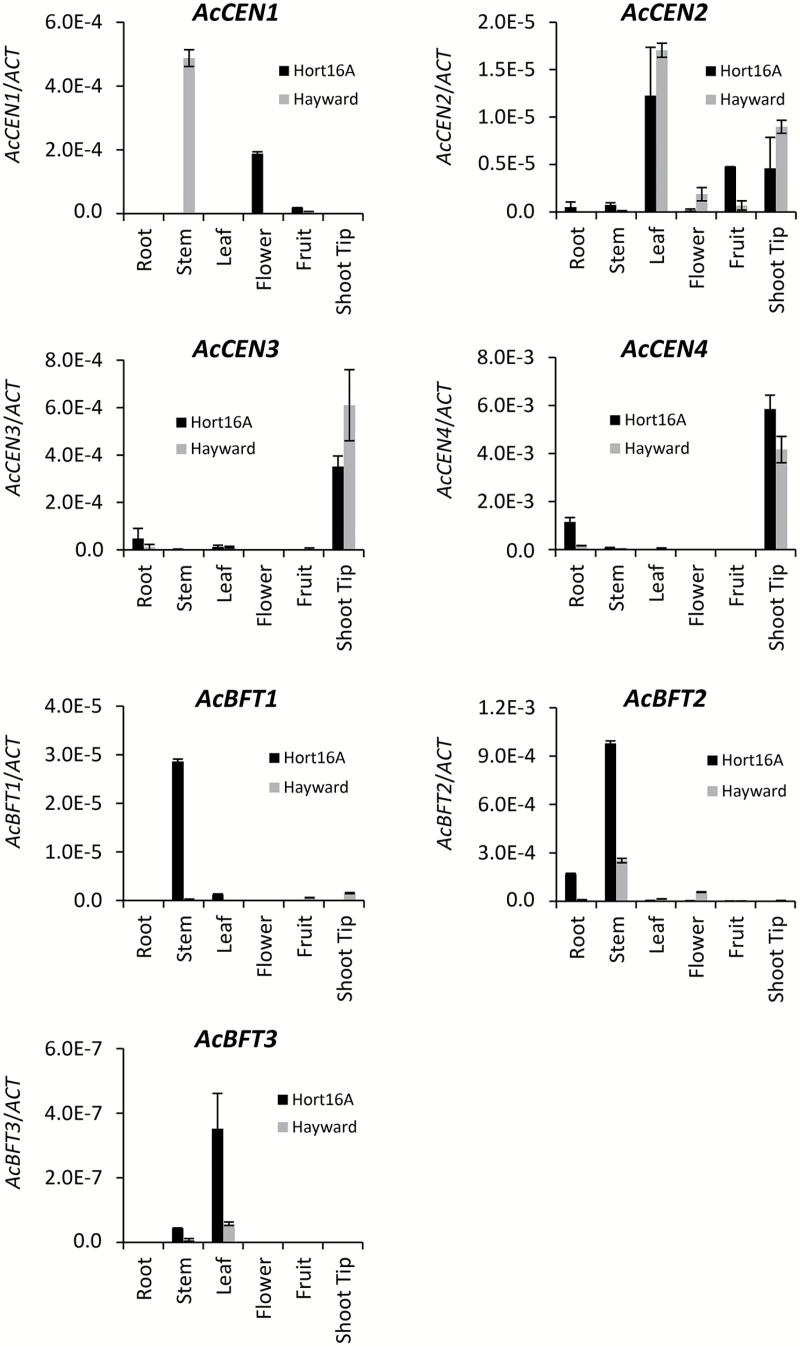
Amplification of the full-length coding sequence of Actinidia PEBP genes often required cDNA from specific tissues (Supplementary Table S2), suggesting differential expression and mostly low expression levels. Real-time PCR amplification using gene-specific oligonucleotide primers was deployed to study organ-specific expression profiles further. In general, low expression was detected for all transcripts (Fig. 2). AcCEN3 and AcCEN4 transcripts were mostly confined to shoot tips, exhibiting a pattern similar to previously described AcCEN (Varkonyi-Gasic et al., 2013). AcCEN2 was detected in all samples, while AcCEN1, phylogenetically most similar to Arabidopsis TFL1 (Fig. 1B), was absent from the shoot tip. All three AcBFT transcripts were amplified from the leaf sample, but AcBFT2 was more abundant in the stem (Fig. 2).
Fig. 2.
Relative expression ±SE of Actinidia CEN- and BFT-like genes in mature field-grown A. chinensis ‘Hort16A’ (black rectangles) and A. deliciosa ‘Hayward’ (gray rectangles) tissues, normalized against ACTIN.
Actinidia TFL1/CEN- and BFT-like genes are functionally conserved floral repressors
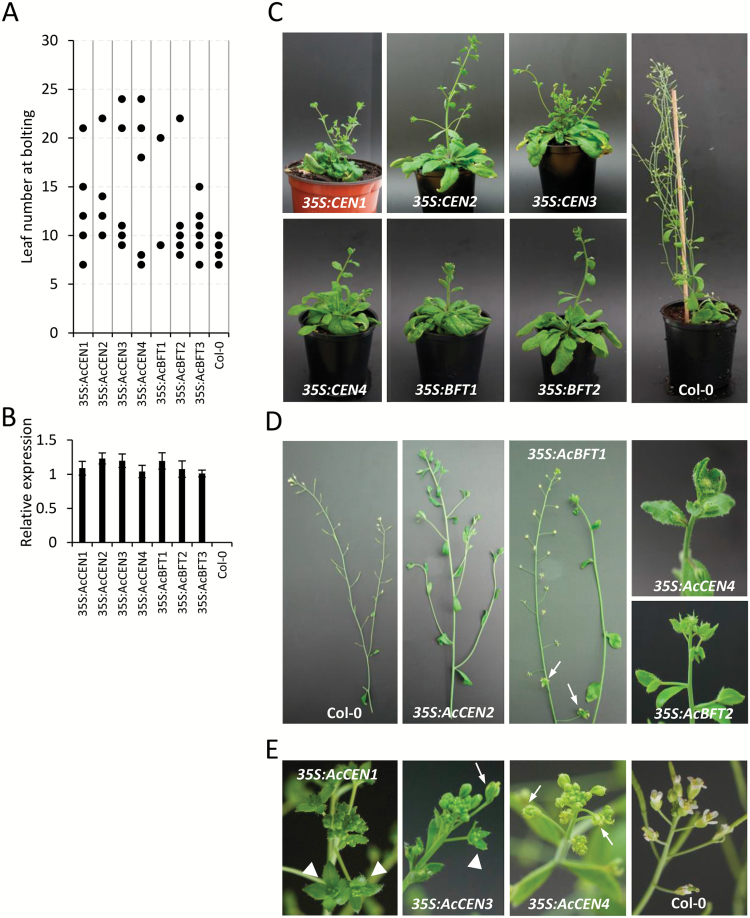
Typically, members of the PEBP family from various flowering plants have conserved roles in the regulation of flowering, so we elected to characterize the kiwifruit PEBP genes by constitutive expression in Arabidopsis. Coding sequences of AcCEN1, AcCEN2, AcCEN3, AcCEN4, AcBFT1, AcBFT2, and AcBFT3 driven by the CaMV 35S promoter were introduced into Arabidopsis Col-0, and a minimum of five kanamycin-resistant primary transgenic plants for each construct were monitored for flowering time in inductive LD conditions. Delayed flowering was observed with all constructs (Fig. 3A), which was correlated with high transgene expression levels (Fig. 3B). A severe delay in bolting combined with change in inflorescence architecture was demonstrated in at least one line for all constructs, with the exception of AcBFT3 which resulted in moderately delayed bolting only. Transgenic plant phenotypes included large rosette leaves, highly branched leafy shoots resulting from proliferation of coflorescences in place of flowers on primary inflorescence stems, leafy axillary shoots developing from axillary meristems of rosette leaves, and third and fourth order branching from the first formed axillary shoots (Fig. 3C, D). The most severe impact was observed in AcCEN4 and AcBFT2 lines, which demonstrated prolonged indeterminacy of the inflorescence meristem (Fig. 3D), and some lines failed to produce flowers and siliques over the duration of the experiment. Development of inflorescences with whorled phyllotaxis of leaf-like organs and flowers with abnormalities such as reduced size, lack of petals, and leaf-like sepals were frequently found (Fig. 3E).
Fig. 3.
Constitutive expression of Actinidia CEN- and BFT-like genes affects flowering in Arabidopsis. (A) The range of flowering time recorded in T1 lines grown in inductive LD conditions. Five to eight lines per construct were monitored for flowering time, recorded as the number of rosette leaves when the primary inflorescence stems were 0.2 cm long. Each dot represents the flowering time of at least one primary transgenic line. (B) Transgene expression in representative late flowering lines, normalized to ACT2. (C) Representative late flowering plants, of the same age and grown under the same conditions as the control Col-0 plant. (D) Examples of altered inflorescence morphology: proliferation of coflorescences on a primary inflorescence stem of a 35S:AcCEN2 plant, leafy axillary shoot produced on a 35S:AcBFT1 plant with abnormal flowers on the primary inflorescence stem, and indeterminate inflorescence meristems of 35S:AcCEN4 and 35S:AcBFT2 plants that failed to produce flowers. Arrows indicate flowers with leaf-like sepals. (E) Examples of inflorescences with whorled phyllotaxis of leaf-like organs (arrowheads) and small abnormal flowers (arrows).
Actinidia MFT-like genes have no impact on flowering time
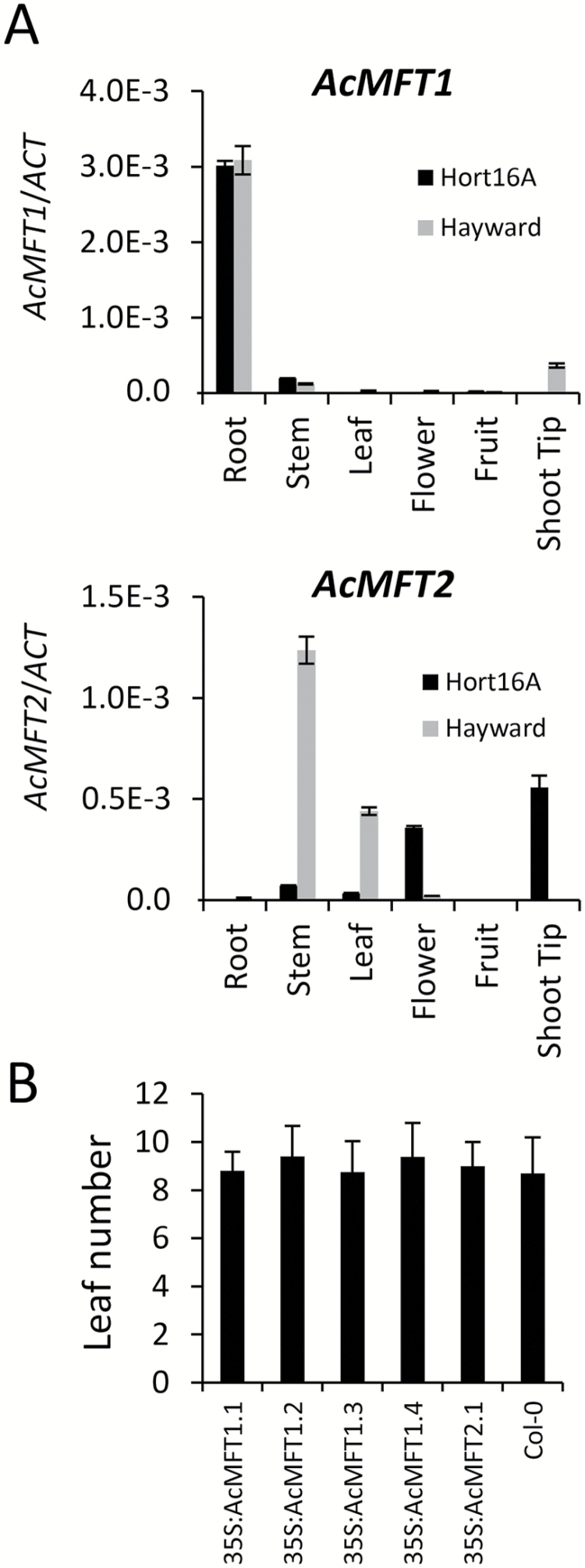
Arabidopsis MFT is highly expressed in developing seeds, but is also detected in root and leaf tissue (Xi et al., 2010), and its constitutive expression led to slightly earlier flowering under LDs (Yoo et al., 2004). Actinidia MFT-like transcripts were abundant in mature fruit and amplified from seed and flower tissue (Supplementary Table S2). Analysis using real-time PCR also identified expression in vegetative tissues for AcMFT2 and particularly prominent root expression for AcMFT1 (Fig. 4A), but very low expression was detected in axillary buds (Supplementary Table S3). To establish if kiwifruit MFT-like genes affect flowering time, coding sequences of AcMFT1 and AcMFT2, each under the control of the 35S promoter, were introduced into Arabidopsis Col-0. Twelve AcMFT1 and four AcMFT2 independent lines were generated and monitored for flowering time in non-inductive SD conditions. None of the plants bolted by the time they had the minimum of 30 rosette leaves. Wild-type flowering time was also observed in their progeny grown in LD conditions (Fig. 4B); therefore, we conclude that kiwifruit MFT-like genes do not promote flowering in transgenic Arabidopsis.
Fig. 4.
Characterization of Actinidia MFT-like genes. (A) Relative expression ±SE in A. chinensis ‘Hort16A’ (black rectangles) and A. deliciosa ‘Hayward’ (gray rectangles) tissues, normalized against ACTIN. (B) Wild-type flowering time in 35S:AcMFT lines. Flowering time was recorded as the number of rosette leaves when the primary inflorescence stems were 0.2 cm long. Error bars represent the SD for 10 T2 transgenic lines grown in long-day (LD) conditions.
Differential and largely opposing expression of AcFT1 and AcFT2
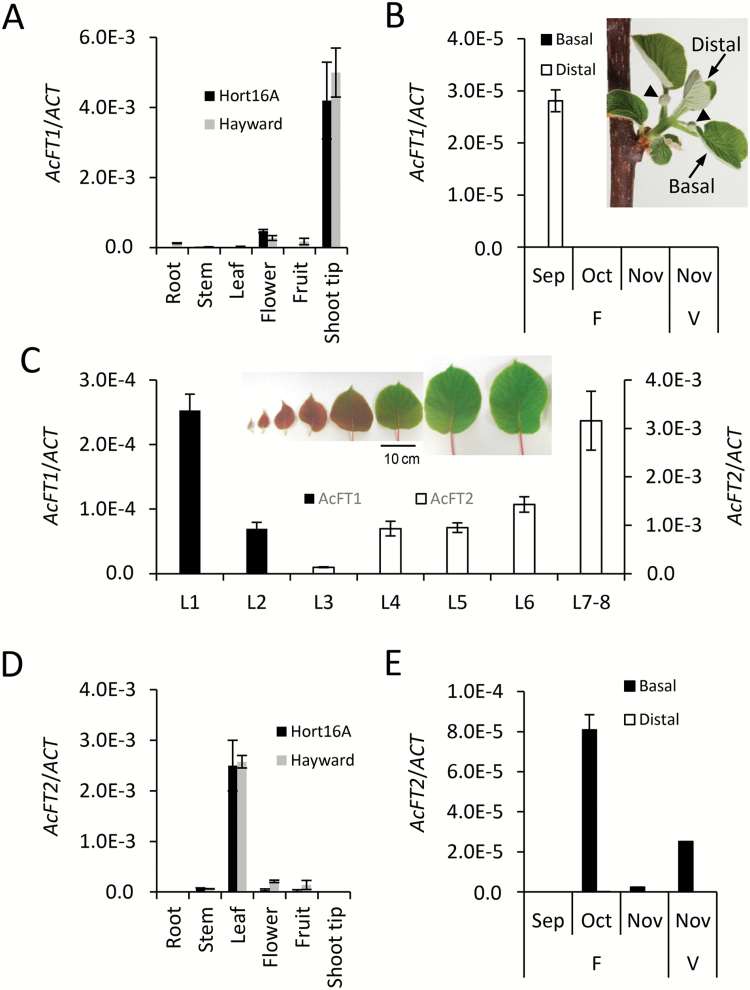
FT-like genes are of particular interest as key activators of flowering and so were studied in detail. AcFT1 was preferentially expressed in terminal buds (Fig. 5A) and in the very small distal leaves during shoot outgrowth in early spring (Fig. 5B). Expression in the shoot tip and very small distal leaves was also observed in rapidly growing large shoots of young glasshouse-grown plants (Fig. 5C; Supplementary Fig. S2). In contrast, AcFT2 was preferentially expressed in the leaf tissue (Fig. 5D). This expression was confined to large basal leaves of larger shoots, declined in mature leaves in late spring, and was detected on branches bearing flower buds, but also on vegetative shoots emerging after the first (floral) flush (Fig. 5E). AcFT2 was also expressed in basal leaves of rapidly growing shoots of young glasshouse-grown plants, with a gradient of expression opposite to that of AcFT1 (Fig. 5C; Supplementary Fig. S2).
Fig. 5.
Expression patterns of AcFT1 and AcFT2 genes. (A) Relative AcFT1 expression ±SE in A. chinensis ‘Hort16A’ (black rectangles) and A. deliciosa ‘Hayward’ (gray rectangles) tissues, normalized against ACTIN. (B) Relative AcFT1 expression in basal and distal leaves of an A. chinensis ‘Hort16A’ growing shoots bearing developing flowers (F) and vegetative shoots (V) during the spring months. The insert shows morphology of a typical A. chinensis shoot in September. (C) Relative expression of AcFT1 and AcFT2 in the leaves of fast growing shoots collected from juvenile glasshouse-grown plants. The insert denotes L1–L8 leaf morphology. Because of the small size of L1 and L2 leaves, leaves collected from single shoots from three plants were pooled for RNA extraction. Another replicate of leaves from three single shoots is presented in Supplementary Fig. S2. (D) Relative AcFT2 expression ±SE in A. chinensis ‘Hort16A’ (black rectangles) and A. deliciosa ‘Hayward’ (gray rectangles) tissues, normalized against ACTIN. (E) Relative AcFT2 expression in basal and distal leaves of A. chinensis ‘Hort16A’ growing shoots bearing flowers (F) and vegetative shoots (V) during spring months. All sampling was performed at mid-day from at least three plants and the expression was normalized against ACTIN.
AcFT1 and AcFT2 are functionally conserved activators of flowering with differential impact on plant morphology
To assess the potential of AcFT1 and AcFT2 to promote flowering, their coding sequences, driven by the 35S promoter, were introduced into the Arabidopsis wild-type Col-0, and eight independent lines for each construct were monitored for flowering time. Early flowering was detected with all AcFT1 and AcFT2 plants in inductive LD conditions and with kanamycin-resistant progeny of two independent lines of each construct in LD and non-inductive SD conditions (Fig. 6A, B). Both constructs were evaluated for functional conservation in the Ler ft-1 late flowering mutant background. Both 35S:AcFT1 and 35S:AcFT2 complemented the ft-1 mutation, and transgenic plants flowered much earlier than wild-type Ler plants (Fig. 6C, D). To test the potential of AcFT2 to perform the role of the leaf-derived phloem-mobile florigen, its coding sequence was placed under control of the vascular-specific SUCROSE TRANSPORTER 2 (SUC2) promoter and introduced into transgenic Arabidopsis. The kanamycin-resistant lines demonstrated precocity in both day-length conditions (Fig. 6A, E). Similarly, SUC2 promoter-driven AcFT1 promoted flowering in transgenic Arabidopsis (Supplementary Fig. S3).
Fig. 6.
Expression of AcFT1 and AcFT2 genes promotes flowering in Arabidopsis. (A) Flowering time in inductive long-day (LD) and non-inductive short-day (SD) conditions. Flowering time was recorded as the number of rosette leaves when the primary inflorescence stems were 0.2 cm long. Error bars represent the standard deviation for 8 each 35S:AcFT1 and 35S:AcFT2 T1 transgenic lines grown in LD conditions, 10 progeny of two T1 lines for each construct grown in LD and SD conditions and 20 SUC2:AcFT2 T1 transgenic lines grown in LD and SD conditions. (B) Differential impact of 35S:AcFT1 and 35S:AcFT2 on leaf size. Plants are of the same age and grown under the same conditions as the control Col-0 plant. (C) Flowering time of the Arabidopsis Ler ft-1 mutant in LD conditions. Flowering time was recorded as the number of rosette leaves when the primary inflorescence stems were 0.2 cm long. Error bars represent the standard deviation for 10 progeny of two T1 lines for each construct grown in LD conditions. (D) Representative early flowering ft1 mutant plants expressing AcFT1 and AcFT2. (E) Plants expressing SUC2:AcFT2 flowered early in LD and SD conditions after normal leaf development. Arrowheads indicate flowers and arrows developing siliques.
35S:AcFT2 Arabidopsis lines typically flowered slightly earlier and often developed terminal flowers, very small leaves, and thin inflorescence stems, in contrast to mostly wild-type size leaves in early-flowering Col-0 35S:AcFT1 lines (Fig. 6B). We hypothesized that AcFT1 divergence in one of the three previously identified positions (G137R in the external loop and I117F or C164G at positions mostly conserved across FT-like proteins; Supplementary Fig. S1) and altered binding to FD contribute to differential FT activity. However, similar phenotypes were observed when we introduced individual mutations at these positions in AcFT2 and reversed them in AcFT1, and lines with simultaneous mutations at all three positions in AcFT2 also produced relatively small rosette leaves (Supplementary Fig. S4). Similarly, both AcFT1 and AcFT2 were capable of interaction with Arabidopsis and kiwifruit FD proteins in a yeast-two hybrid assay, with similar interaction intensities (Supplementary Fig. S5).
Constitutive expression of AcFT1 and AcFT2 leads to extreme precocity in transgenic kiwifruit
To test if AcFT1 and AcFT2 promote flowering and demonstrate differential activity in kiwifruit, their coding sequences driven by the 35S promoter were introduced into A. chinensis using standard regeneration and transformation protocols. Major differences were observed 2 months after Agrobacterium inoculation; flower instead of leaf development was observed in the presence of ectopic AcFT2, but both leaf and flower development were recorded in the presence of ectopic AcFT1 and only leaf development was recorded in the control (Fig. 7A). Both constructs gave rise to in vitro flowers (Fig. 7B, C), which aborted development soon after initiation of rudimentary reproductive organs (Fig. 7D), while some AcFT1 lines developed leaves and flowers with differentiating floral organs (Fig. 7E).
Fig. 7.
Transgenic kiwifruit flowering in vitro. (A) Flower and leaf, only flower, and only leaf develop in 35S:AcFT1, 35S:AcFT2, and control lines, respectively, 2 months after Agrobacterium inoculation. (B, C) In vitro flowers with undifferentiated floral organs. (D) Cross-section of an in vitro flower. The arrow indicates a rudimentary anther. (E) Leaves and differentiating floral organs in 35S:AcFT1 lines. Scale bars=10 mm (A) and 1 mm (B–E).
Seasonal expression patterns in axillary buds
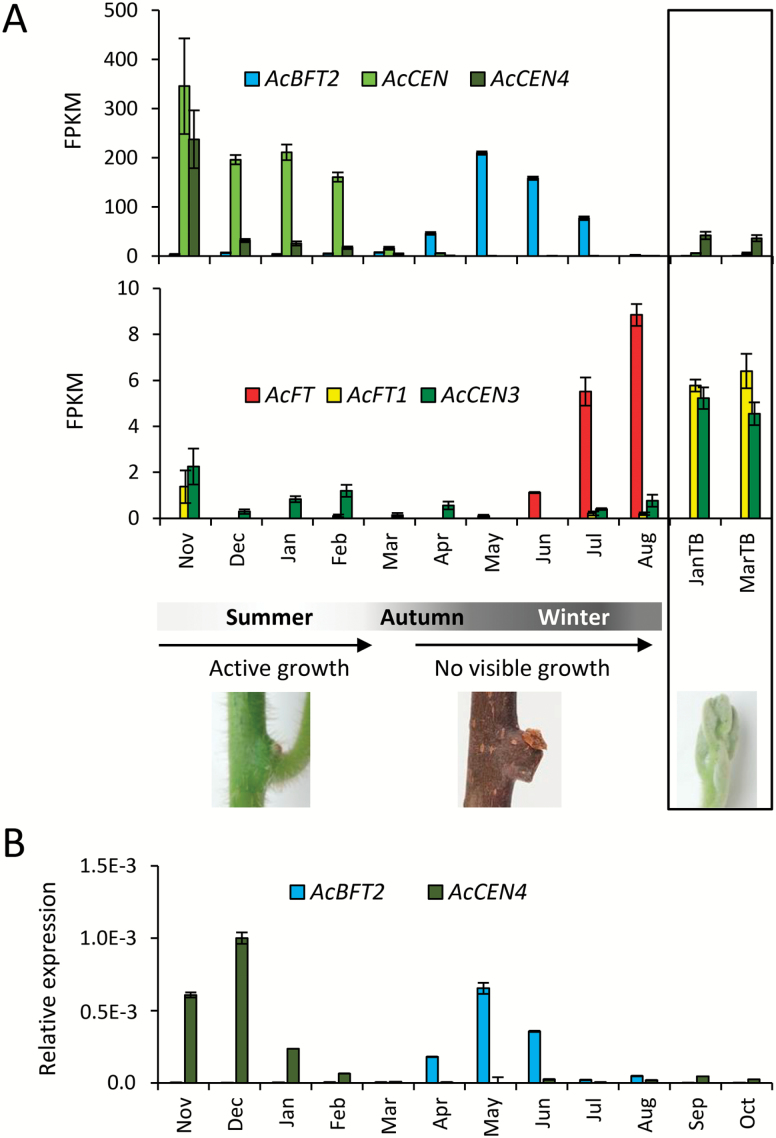
To study the potential roles in dormancy, winter chilling, growth resumption, and initiation of flowering, expression of PEBP genes was further analyzed in kiwifruit axillary buds collected at regular intervals over the season. Interrogation of axillary bud transcriptomes identified differential expression of some Actinidia CEN- and BFT-like genes at different stages of bud development. AcCEN and AcCEN4 were abundant in summer, during active growth, but gradually declined at the beginning of autumn, at which stage AcBFT2 transcript abundance started to increase and remained high during the dormancy period. AcCEN4 and to a lesser extent AcCEN and AcCEN3 were also detected in actively growing terminal buds collected in summer and early autumn (Fig. 8A). Very low abundance or total absence of transcripts was observed for the remaining kiwifruit CEN- and BFT-like genes (Supplementary Table S3). Unlike AcFT, which was detected in buds during accumulation of winter chilling and growth resumption in spring, AcFT1 and AcFT2 transcripts were mainly absent from axillary buds and AcFT1 was detected in samples of actively growing terminal buds collected in summer and early autumn (Fig. 8A). Accumulation of AcBFT2 during establishment of dormancy was also detected in ‘Hayward’ axillary buds (Fig. 8B). Transcriptional activity of expressed PEBP genes is summarized in a schematic diagram (Fig. 9).
Fig. 8.
Expression in buds during the season. (A) A. chinensis axillary buds were collected at monthly intervals during the growth and dormancy cycle, and subjected to RNA-seq. Only uniquely mapped reads with no mismatches per paired alignment were used for gene expression studies. Frequency of gene expression was calculated as fragments per kilobase of transcript per million reads (FPKM) and is presented as the average ±SE of three biological replicates. The seasons and appearance of axillary buds are indicated below. Box, actively growing terminal buds were collected in January and March and subjected to RNA-seq as above. Frequency of gene expression is presented as the average ±SE of three and two biological replicates for January and March samples, respectively. The appearance of actively growing terminal buds is indicated below. (B) Relative expression ±SE in A. deliciosa axillary buds collected at monthly intervals during the growth and dormancy cycle, normalized against ACTIN. Expression of previously described genes is presented in Supplementary Fig. S6.
Fig. 9.
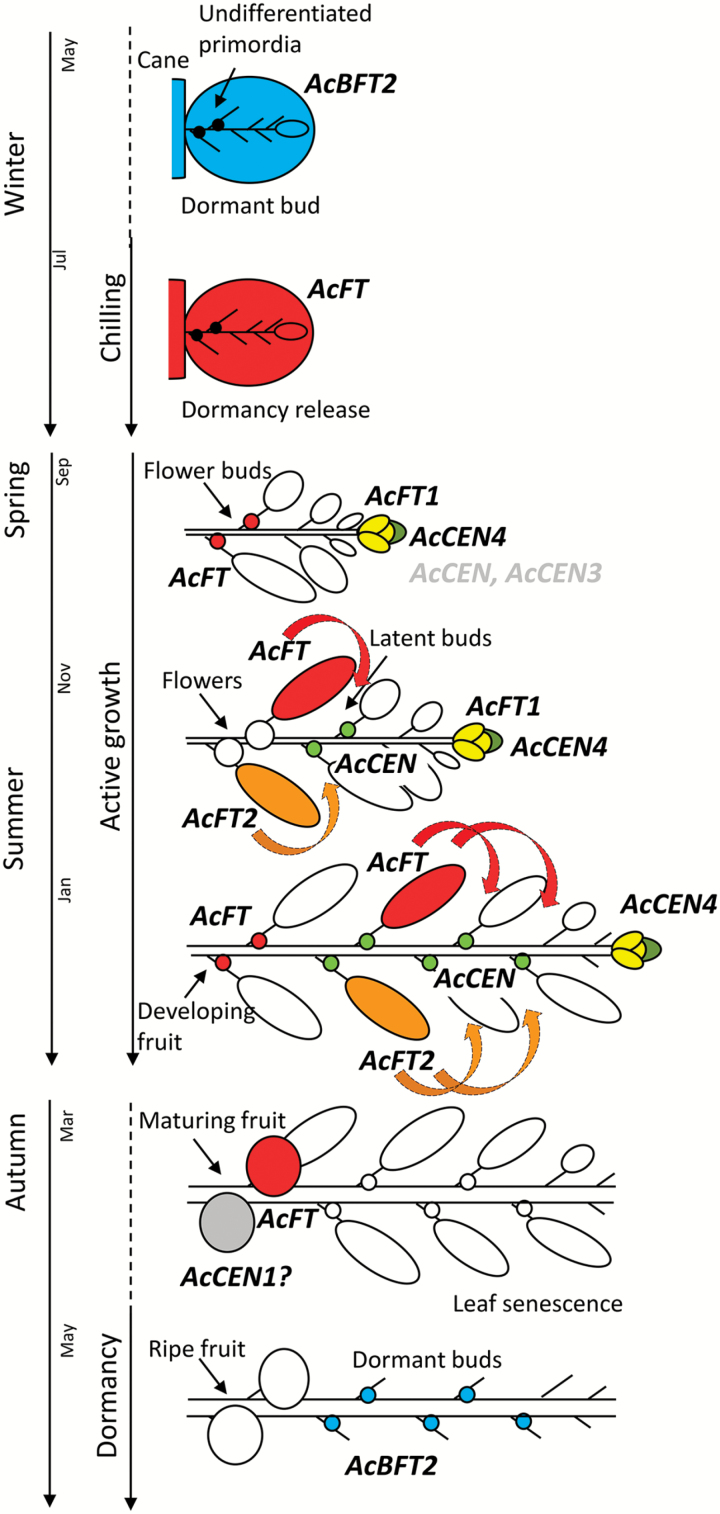
Schematic summarizing accumulation of transcripts in the shoot bud and developing shoot. AcBFT2 (blue) accumulates at growth cessation and during dormancy in the bud. Undifferentiated meristems in the axils of leaf primordia are indicated by black dots. AcFT (red) accumulates with winter chilling and dormancy release. Shoot outgrowth is marked by expression of AcCEN4 (dark green) and to a lesser degree AcCEN and AcCEN3, accompanied by accumulation of AcFT1 (yellow). Subsequently, AcFT2 (orange) accumulates in large, basal leaves. This expression is dynamic, declines with maturation, and is similar to that reported for AcFT. Dynamic accumulation of AcFT during flower and fruit development, but decline at maturation was also described (Varkonyi-Gasic et al., 2013). AcCEN1 (gray) is potentially associated with fruit development. AcCEN (light green) and to a lesser degree AcCEN4 accumulate in developing (latent) buds during active shoot growth. Subsequently, AcCEN declines and AcBFT accumulates with the onset of dormancy. Potential FT protein movement is indicated by colored curved arrows.
Discussion
The expansion and diversification of Actinidia floral repressor genes
Evolution of the PEBP family has had an important role in plant diversification and adaptation. Actinidia have undergone two recent whole-genome duplication events, which followed the ancient hexaploidization event shared by core eudicots (Huang et al., 2013). As a result of these events, the Actinidia PEBP gene family contains multiple FT-, CEN-, and BFT-like genes. The expanded family of floral repressors in kiwifruit might be of significant relevance to plant growth, architecture, and flowering. All kiwifruit CEN- and BFT-like genes drive indeterminacy and delay flowering in Arabidopsis, and at least one of them, AcCEN, inhibits flowering in Actinidia eriantha (Varkonyi-Gasic et al., 2013). Together with AcCEN, AcCEN3 and AcCEN4 appear to be preferentially expressed in the buds and are thus likely to perform a role in regulation of flowering, architecture, and determinacy as functional homologs of Arabidopsis TFL1. Determinacy is an agronomically important trait associated with domestication, and homologs of TFL1/CEN genes have been implicated in agricultural adaptation in crops such as tomato, barley, grapevine, bean, soybean, and strawberry (Pnueli et al., 1998; Liu et al., 2010; Fernandez et al., 2010; Tian et al., 2010; Comadran et al., 2012; Iwata et al., 2012; Koskela et al., 2012; Repinski et al., 2012). Kiwifruit has been developed into a horticultural crop only recently (Ferguson, 1990). Most cultivars are either direct selections from the wild or are only a few generations removed from the wild, with only a short history of systematic breeding (Datson and Ferguson, 2011). Currently, excessive vegetative vigor is managed through costly manual pruning. For that reason, combined with generally low harvest index of current kiwifruit cultivars, manipulation of the shoot architecture remains a significant breeding target. Therefore, AcCEN, AcCEN4, and AcCEN3 represent prime candidates for kiwifruit improvement.
In contrast to kiwifruit CEN-like transcripts, which were associated with active growth and declined before the establishment of dormancy, expression of a member of the kiwifruit BFT clade is highest during winter dormancy and peaks at the time of leaf drop. The role of BFT proteins remains poorly understood. Arabidopsis BFT has a TFL1-like activity (Yoo et al., 2010) and is associated with stress response (Ryu et al., 2011, 2014). Up-regulation of Arabidopsis BFT following abscisic acid (ABA), drought, or osmotic stress treatment (Chung et al., 2010) resembles AcBFT2 transcript accumulation during winter dormancy, a process which is also potentially mediated by ABA and involves cold-, drought-, and stress-regulated genes (Horvath et al., 2008). In Arabidopsis, high BFT expression causes suppression of axillary inflorescence development (Yoo et al., 2010). In kiwifruit, AcBFT2 transcript accumulation is associated with suppression of latent bud growth during unfavorable winter conditions. Thus, AcBFT2 may repress vegetative growth, but could also maintain the undifferentiated state of meristems in the axils of leaf primordia in the dormant bud (Fig. 9), which develop as inflorescences after sufficient chilling. Interestingly, apple MdBFTa and MdBFTb were expressed at higher levels in dwarfing rootstocks, commonly used to reduce the scion vigor, suggesting a conserved role in suppression of axillary shoot development (Foster et al., 2013). Similarly, expression of a grapevine BFT homolog, VvTFL1C, increased progressively during latent bud development (Carmona et al., 2007), suggesting that aspects of BFT function may be conserved in woody perennials.
AcCEN1, phylogenetically closest to TFL1, and AcCEN2 appear to be expressed in a range of tissues, including the fruit. The relevance of this is unclear at this stage, but is in line with reports of TFL1/CEN-like gene expression in fleshy fruit (Lindqvist et al., 2006; Mimida et al., 2009; Li et al., 2014). The current study provides no evidence that MFT-like genes contribute towards regulation of flowering and architecture in kiwifruit. Their expression in seed may be similar to a proposed role in hormonal signaling regulating seed germination in Arabidopsis (Xi et al., 2010; Footitt et al., 2011) and wheat (Nakamura et al., 2011), but the universality of this MFT-like role is yet to be confirmed in kiwifruit.
The diversification of Actinidia FT genes
The FT-like genes have emerged with the evolution of the angiosperms, consistent with their key role in regulation of flowering (Shalit et al., 2009; Karlgren et al., 2011), while subsequent functional divergence contributed to the redundant, similar, or novel, and in some cases antagonistic FT functions (Blackman et al., 2010; Pin et al., 2010; Hsu et al., 2011;Navarro, 2011; Harig et al., 2012; Lee et al., 2013; Cao et al., 2016). The three Actinidia FT genes probably reflect two recent whole-genome duplication events in kiwifruit (Huang et al., 2013), after which these genes have evolved divergent expression patterns. The previously described AcFT gene expression in response to cold correlated with winter chilling requirement and bloom time of kiwifruit cultivars, but it failed to promote flowering in kiwifruit (Varkonyi-Gasic et al., 2013). This expression at later stages of dormancy coincided with reduction in AcBFT2 transcript accumulation, suggesting that antagonism between these two genes may be of importance for bud dormancy and growth cycles.
In contrast, AcFT1 and AcFT2 both act as potent activators of flowering. This function is conserved across diverse plant species and may be non-cell autonomous, as demonstrated for AcFT2, thus fulfilling the criteria for florigen (Chailakhyan, 1968). The severity of phenotypes in both Arabidopsis and kiwifruit plants constitutively expressing high levels of AcFT2 suggests that it may act in a dosage-dependent manner across long distances, when high expression may provide sufficient amounts to be delivered to the meristems in latent buds, to balance out the repressors of flowering and establish floral fate. Conversely, AcFT1 expression differs from the florigen concept in that its expression appears restricted to kiwifruit sink tissue. Therefore, it might be fulfilling a role other than flowering control, perhaps in regulation of terminal bud expansion rate and abortion, which underlies high developmental plasticity of kiwifruit (Foster et al., 2007). Alternatively, both AcFT1 and AcFT2 may act locally; in particular, AcFT2 may have a role in termination of leaf expansion and development, while AcFT1 may regulate termination of the shoot apical meristem after establishment of leaf fate.
Comparison of amino acid sequences identified divergence of AcFT1 residues in positions highly conserved in other FT-like proteins, which did not abolish its function and had no effect on the interaction with FD proteins, further confirming the robust nature of FT proteins as activators of the flowering process. The substitution for polar-positive arginine in the position of non-polar-neutral glycine in the conserved segment B (G137R) had little effect on AcFT1 activity, as recently demonstrated in Arabidopsis (Ho and Weigel, 2014) and consistent with findings in sugar beet (Klintenäs et al., 2012). A conservative I117F substitution did not affect FT function, despite conservation of isoleucine in this position in FT and most other PEBP proteins (Supplementary Fig. S1) (Ho and Weigel, 2014). Similarly, introduction of a glycine in the position of highly conserved cysteine (C164G) had no impact on AcFT activity. Therefore, the sum of differences in these and other positions and interactions with proteins other than FD, including TEOSINTE-BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors (Mimida et al., 2011; Niwa et al., 2013; Ho and Weigel, 2014), and binding of unknown ligand(s) might provide the basis for different flowering time and architecture of 35S:AcFT1 and 35S:AcFT2 lines.
In summary, multiple differentially expressed and functionally divergent FT- and CEN-like genes regulate flowering and architecture in kiwifruit, and a BFT-like gene is associated with a lack of growth during dormancy. Specific expression of FT-like activators and CEN/BFT-like repressors (Fig. 9) may ensure adequate timing of growth and dormancy, leaf, flower, and fruit development, shoot tip growth, and establishment of latent buds. Manipulation of their dosage and functionality may provide practical means to reduce juvenility, hasten the breeding process, improve orchard management, and increase kiwifruit productivity.
Supplementary Material
Acknowledgements
We thank Monica Dragulescu for assistance with transgenic plants, Tim Holmes and Minna Pesonen for assistance with photography, and Anne Gunson for critically reading the manuscript. The Arabidopsis SUC2 promoter was kindly provided by Dr Ruth Stadler, and the Arabidopsis ft-1 mutant was obtained from the European Arabidopsis Stock Centre. The authors declare no conflict of interest. This work was funded by the New Zealand Ministry of Business, Innovation and Employment grants C10X0816 and C11X1602.
References
- Abe M, Kobayashi Y, Yamamoto S, et al. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. 2006. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO Journal 25, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH. 2010. The role of recently derived FT paralogs in sunflower domestication. Current Biology 20, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. 2006. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. 1996. Control of inflorescence architecture in Antirrhinum. Nature 379, 791–797. [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. 1997. Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. [DOI] [PubMed] [Google Scholar]
- Brundell D. 1975a Flower development of the Chinese gooseberry (Actinidia chinensis Planch.). I. Development of the flower shoot. New Zealand Journal of Botany 13, 473–483. [Google Scholar]
- Brundell D. 1975b Flower development of the Chinese goosberry (Actinidia chinensis Planch.). II. Development of the flowering bud. New Zealand Journal of Botany 13, 485–496. [Google Scholar]
- Brundell DJ. 1976. The effect of chilling on the termination of rest and flower bud development of the chinese goosberry. Scientia Horticulturae 4, 175–182. [Google Scholar]
- Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S. 2016. Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Frontiers in Plant Science 6, 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona MJ, Calonje M, Martínez-Zapater JM. 2007. The FT/TFL1 gene family in grapevine. Plant Molecular Biology 63, 637–650. [DOI] [PubMed] [Google Scholar]
- Chailakhyan MK. 1968. Internal factors of plant flowering. Annual Review of Plant Physiology 19, 1–37. [Google Scholar]
- Chung KS, Yoo SY, Yoo SJ, Lee JS, Ahn JH. 2010. BROTHER OF FT AND TFL1 (BFT), a member of the FT/TFL1 family, shows distinct pattern of expression during the vegetative growth of Arabidopsis. Plant Signaling and Behavior 5, 1102–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, et al. 2012. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nature Genetics 44, 1388–1392. [DOI] [PubMed] [Google Scholar]
- Conti L, Bradley D. 2007. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. The Plant Cell 19, 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, et al. 2008. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics 9, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Ananiev EV. 2010. Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiology 153, 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson PM, Ferguson AR. 2011. Actinidia. In: Kole C, ed. Wild crop relatives: genomic and breeding resources. Berlin: Springer, 1–20. [Google Scholar]
- Ding J, Nilsson O. 2016. Molecular regulation of phenology in trees—because the seasons they are a-changin’. Current Opinion in Plant Biology 29, 73–79. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RS, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC. 2009. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiology 151, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Shimada T, Fujii H, Kobayashi Y, Araki T, Omura M. 2005. Ectopic expression of an FT homolog from citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Research 14, 703–712. [DOI] [PubMed] [Google Scholar]
- Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K. 2015. Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Scientific Reports 5, 12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson A. 1990. The genus Actinidia. In: Warrington IJ, Weston GC, eds. Kiwifruit: science and management. Auckland: Ray Richards Publisher in association with the New Zealand Society for Horticultural Science, 15–35. [Google Scholar]
- Fernandez L, Torregrosa L, Segura V, Bouquet A, Martinez-Zapater JM. 2010. Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. The Plant Journal 61, 545–557. [DOI] [PubMed] [Google Scholar]
- Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE. 2011. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proceedings of the National Academy of Sciences, USA 108, 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TM, Seleznyova AN, Barnett AM. 2007. Independent control of organogenesis and shoot tip abortion are key factors to developmental plasticity in kiwifruit (Actinidia). Annals of Botany 100, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TM, Watson AE, Hooijdonk BM, Schaffer RJ. 2013. Key flowering genes including FT-like genes are upregulated in the vasculature of apple dwarfing rootstocks. Tree Genetics and Genomes 10, 189–202. [Google Scholar]
- Freiman A, Golobovitch S, Yablovitz Z, et al. 2015. Expression of flowering locus T2 transgene from Pyrus communis L. delays dormancy and leaf senescence in Malus×domestica Borkh, and causes early flowering in tobacco. Plant Science 241, 164–176. [DOI] [PubMed] [Google Scholar]
- Freiman A, Shlizerman L, Golobovitch S, et al. 2012. Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2. Planta 235, 1239–1251. [DOI] [PubMed] [Google Scholar]
- Haberman A, Ackerman M, Crane O, Kelner JJ, Costes E, Samach A. 2016. Different flowering response to various fruit loads in apple cultivars correlates with degree of transcript reaccumulation of a TFL1-encoding gene. The Plant Journal 87, 161–173. [DOI] [PubMed] [Google Scholar]
- Hanano S, Goto K. 2011. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. The Plant Cell 23, 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D. 2005. A single amino acid converts a repressor to an activator of flowering. Proceedings of the National Academy of Sciences, USA 102, 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harig L, Beinecke FA, Oltmanns J, et al. 2012. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. The Plant Journal 72, 908–921. [DOI] [PubMed] [Google Scholar]
- Hedman H, Källman T, Lagercrantz U. 2009. Early evolution of the MFT-like gene family in plants. Plant Molecular Biology 70, 359–369. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WW, Weigel D. 2014. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. The Plant Cell 26, 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson JV. 2008. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C-Y, Adams JP, Kim H, et al. 2011. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proceedings of the National Academy of Sciences, USA 108, 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Liu Y, Luthe DS, Yuceer C. 2006. Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. The Plant Cell 18, 1846–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ding J, Deng D, et al. 2013. Draft genome of the kiwifruit Actinidia chinensis. Nature Communications 4, 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Gaston A, Remay A, et al. 2012. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. The Plant Journal 69, 116–125. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Müller AE. 2009. Flowering time control and applications in plant breeding. Trends in Plant Science 14, 563–573. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Karlgren A, Gyllenstrand N, Källman T, Sundström JF, Moore D, Lascoux M, Lagercrantz U. 2011. Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiology 156, 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klintenäs M, Pin PA, Benlloch R, Ingvarsson PK, Nilsson O. 2012. Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytologist 196, 1260–1273. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koskela EA, Mouhu K, Albani MC, et al. 2012. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiology 159, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela EA, Sønsteby A, Flachowsky H, Heide OM, Hanke MV, Elomaa P, Hytönen T. 2016. TERMINAL FLOWER1 is a breeding target for a novel everbearing trait and tailored flowering responses in cultivated strawberry (Fragaria × ananassa Duch.). Plant Biotechnology Journal 14, 1852–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoda N, Hayashi H, Suzuki M, et al. 2010. Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus×domestica Borkh.). Plant and Cell Physiology 51, 561–575. [DOI] [PubMed] [Google Scholar]
- Kotoda N, Iwanami H, Takahashi S, Abe K. 2006. Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. Journal of the American Society for Horticultural Science 131, 74–81. [Google Scholar]
- Ledger SE, Janssen BJ, Karunairetnam S, Wang T, Snowden KC. 2010. Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytologist 188, 803–813. [DOI] [PubMed] [Google Scholar]
- Lee R, Baldwin S, Kenel F, McCallum J, Macknight R. 2013. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nature Communications 4, 2884. [DOI] [PubMed] [Google Scholar]
- Li C, Luo L, Fu Q, Niu L, Xu Z-F. 2014. Identification and characterization of the FT/TFL1 gene family in the biofuel plant Jatropha curcas. Plant Molecular Biology Reporter, 1–8. [Google Scholar]
- Lifschitz E, Ayre BG, Eshed Y. 2014. Florigen and anti-florigen—a systemic mechanism for coordinating growth and termination in flowering plants. Frontiers in Plant Science 5, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist C, Scheen AC, Yoo MJ, et al. 2006. An expressed sequence tag (EST) library from developing fruits of an Hawaiian endemic mint (Stenogyne rugosa, Lamiaceae): characterization and microsatellite markers. BMC Plant Biology 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Watanabe S, Uchiyama T, et al. 2010. The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiology 153, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Trujillo M, Limones-Briones V, Cabrera-Ponce JL, Herrera-Estrella L. 2004. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Molecular Biology Reporter 22, 63–70. [Google Scholar]
- Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W. 2001. Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes to Cells 6, 327–336. [DOI] [PubMed] [Google Scholar]
- Mimida N, Kidou S, Iwanami H, Moriya S, Abe K, Voogd C, Varkonyi-Gasic E, Kotoda N. 2011. Apple FLOWERING LOCUS T proteins interact with transcription factors implicated in cell growth and organ development. Tree Physiology 31, 555–566. [DOI] [PubMed] [Google Scholar]
- Mimida N, Kotoda N, Ueda T, Igarashi M, Hatsuyama Y, Iwanami H, Moriya S, Abe K. 2009. Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus×domestica Borkh.). Plant and Cell Physiology 50, 394–412. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Abe F, Kawahigashi H, et al. 2011. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. The Plant Cell 23, 3215–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, Shimamoto K, Prat S. 2011. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. [DOI] [PubMed] [Google Scholar]
- Nishikawa F, Endo T, Shimada T, Fujii H, Shimizu T, Omura M, Ikoma Y. 2007. Increased CiFT abundance in the stem correlates with floral induction by low temperature in Satsuma mandarin (Citrus unshiu Marc.). Journal of Experimental Botany 58, 3915–3927. [DOI] [PubMed] [Google Scholar]
- Niwa M, Daimon Y, Kurotani K, et al. 2013. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. The Plant Cell 25, 1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F. 1997. Cloning and molecular analysis of the Arabidopsis gene TERMINAL FLOWER 1. Molecular and General Genetics 254, 186–194. [DOI] [PubMed] [Google Scholar]
- Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O. 2010. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400. [DOI] [PubMed] [Google Scholar]
- Pin PA, Nilsson O. 2012. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant, Cell and Environment 35, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. 1998. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. 2001. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. The Plant Cell 13, 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Varkonyi-Gasic E. 2016. FT and florigen long-distance flowering control in plants. Current Opinion in Plant Biology 33, 77–82. [DOI] [PubMed] [Google Scholar]
- Putterill J, Zhang L, Yeoh CC, Balcerowicz M, Jaudal M, Varkonyi-Gasic E. 2013. FT genes and regulation of flowering in the legume Medicago truncatula. Functional Plant Biology 40, 1199–1207. [DOI] [PubMed] [Google Scholar]
- Randoux M, Davière JM, Jeauffre J, et al. 2014. RoKSN, a floral repressor, forms protein complexes with RoFD and RoFT to regulate vegetative and reproductive development in rose. New Phytologist 202, 161–173. [DOI] [PubMed] [Google Scholar]
- Rantanen M, Kurokura T, Jiang P, Mouhu K, Hytönen T. 2015. Strawberry homologue of TERMINAL FLOWER1 integrates photoperiod and temperature signals to inhibit flowering. The Plant Journal 82, 163–173. [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ. 1998. A common mechanism controls the life cycle and architecture of plants. Development 125, 1609–1615. [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES. 1999. Separation of shoot and floral identity in Arabidopsis. Development 126, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Repinski SL, Kwak M, Gepts P. 2012. The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theoretical and Applied Genetics 124, 1539–1547. [DOI] [PubMed] [Google Scholar]
- Rinne PL, Welling A, Vahala J, Ripel L, Ruonala R, Kangasjärvi J, van der Schoot C. 2011. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. The Plant Cell 23, 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JY, Lee HJ, Seo PJ, Jung JH, Ahn JH, Park CM. 2014. The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Molecular Plant 7, 377–387. [DOI] [PubMed] [Google Scholar]
- Ryu JY, Park CM, Seo PJ. 2011. The floral repressor BROTHER OF FT AND TFL1 (BFT) modulates flowering initiation under high salinity in Arabidopsis. Molecules and Cells 32, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, Lifschitz E. 2009. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proceedings of the National Academy of Sciences, USA 106, 8392–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR. 1991. A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. The Plant Cell 3, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelgar WP, Clearwater MJ, Walton EF. 2007. Flowering of kiwifruit (Actinidia deliciosa) is reduced by long photoperiods. New Zealand Journal of Crop and Horticultural Science 35, 33–38. [Google Scholar]
- Song GQ, Walworth A, Zhao D, Jiang N, Hancock JF. 2013. The Vaccinium corymbosum FLOWERING LOCUS T-like gene (VcFT): a flowering activator reverses photoperiodic and chilling requirements in blueberry. Plant Cell Reports 32, 1759–1769. [DOI] [PubMed] [Google Scholar]
- Sreekantan L, Thomas MR. 2006. VvFT and VvMADS8, the grapevine homologues of the floral integrators FT and SOC1, have unique expression patterns in grapevine and hasten flowering in Arabidopsis. Functional Plant Biology 33, 1129–1139. [DOI] [PubMed] [Google Scholar]
- Srinivasan C, Dardick C, Callahan A, Scorza R. 2012. Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS One 7, e40715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J. 2010. Artificial selection for determinate growth habit in soybean. Proceedings of the National Academy of Sciences, USA 107, 8563–8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Moss SM, Voogd C, Wang T, Putterill J, Hellens RP. 2013. Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit. New Phytologist 198, 732–746. [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Moss SM, Voogd C, Wu R, Lough RH, Wang YY, Hellens RP. 2011. Identification and characterization of flowering genes in kiwifruit: sequence conservation and role in kiwifruit flower development. BMC Plant Biology 11, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogd C, Wang T, Varkonyi-Gasic E. 2015. Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. Journal of Experimental Botany 66, 4699–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E, Fowke P, Weis K, McLeay P. 1997. Shoot axillary bud morphogenesis in kiwifruit (Actinidia deliciosa). Annals of Botany 80, 13–21. [Google Scholar]
- Walton EF, Podivinsky E, Wu RM. 2001. Bimodal patterns of floral gene expression over the two seasons that kiwifruit flowers develop. Physiologia Plantarum 111, 396–404. [DOI] [PubMed] [Google Scholar]
- Wang T, Atkinson R, Janssen B. 2007. The choice of Agrobacterium strain for transformation of kiwifruit. ISHS Acta Horticulturae 753, 227–232. [Google Scholar]
- Wenzel S, Flachowsky H, Hanke M-V. 2013. The Fast-track breeding approach can be improved by heat-induced expression of the FLOWERING LOCUS T genes from poplar (Populus trichocarpa) in apple (Malus × domestica Borkh.). Plant Cell, Tissue and Organ Culture 115, 127–137. [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wu RM, Walton EF, Richardson AC, Wood M, Hellens RP, Varkonyi-Gasic E. 2012. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. Journal of Experimental Botany 63, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H. 2010. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. The Plant Cell 22, 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, Kishigami R, Yoshikawa N. 2014. Reduced generation time of apple seedlings to within a year by means of a plant virus vector: a new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnology Journal 12, 60–68. [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Li C, Yoshikawa N. 2016. Promotion of flowering by Apple latent spherical virus vector and virus elimination at high temperature allow accelerated breeding of apple and pear. Frontiers in Plant Science 7, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, Sasaki S, Yamagata K, Komori S, Nagase M, Wada M, Yamamoto T, Yoshikawa N. 2011. Promotion of flowering and reduction of a generation time in apple seedlings by ectopical expression of the Arabidopsis thaliana FT gene using the Apple latent spherical virus vector. Plant Molecular Biology 75, 193–204. [DOI] [PubMed] [Google Scholar]
- Ye J, Geng Y, Zhang B, Mao H, Qu J, Chua N-H. 2014. The Jatropha FT ortholog is a systemic signal regulating growth and flowering time. Biotechnology for Biofuels 7, 1–11.24387051 [Google Scholar]
- Yeoh CC, Balcerowicz M, Laurie R, Macknight R, Putterill J. 2011. Developing a method for customized induction of flowering. BMC Biotechnology 11, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, et al. 1999. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401, 173–177. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Chung KS, Jung SH, Yoo SY, Lee JS, Ahn JH. 2010. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. The Plant Journal 63, 241–253. [DOI] [PubMed] [Google Scholar]
- Yoo SY, Kardailsky I, Lee JS, Weigel D, Ahn JH. 2004. Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Molecules and Cells 17, 95–101. [PubMed] [Google Scholar]
- Zeevaart JA. 2008. Leaf-produced floral signals. Current Opinion in Plant Biology 11, 541–547. [DOI] [PubMed] [Google Scholar]
- Zhang H, Harry DE, Ma C, Yuceer C, Hsu CY, Vikram V, Shevchenko O, Etherington E, Strauss SH. 2010. Precocious flowering in trees: the FLOWERING LOCUS T gene as a research and breeding tool in Populus. Journal of Experimental Botany 61, 2549–2560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.