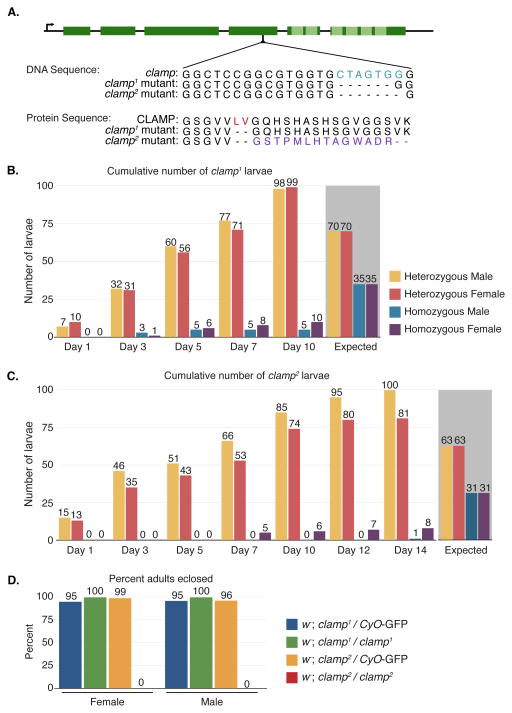
Figure 1. The clamp2 mutation is homozygous lethal and the clamp1 allele is homozygous viable.
(A) The CRISPR/Cas9-introduced frameshift is located in the fourth exon (dark green boxes) of the clamp gene, upstream of the DNA binding domain containing six zinc fingers (light green boxes). The homozygous viable clamp1 mutation is a six base pair deletion (blue), resulting in a deletion of two amino acids (red) and an in-frame shift of the protein sequence. The homozygous lethal clamp2 mutation consists of the same six base pair deletion (blue), with an additional seventh base removed. This causes a frameshift of the protein sequence (purple) resulting in an early termination codon.
(B) The cumulative number of larvae counted for both male and female clamp1 heterozygous and homozygous animals is shown. In total, 212 larvae were counted. Day 1 indicates the first day in which wandering third instar larvae began emerging. Homozygous males (blue) and females (purple) began emerging two days after their heterozygous siblings. The expected number of larvae out of 212 for each sex and genotype is indicated with the grey background.
(C) The cumulative number of larvae counted for both male and female clamp2 heterozygous and homozygous animals. In total, 190 larvae were counted. Day 1 indicates the first day in which wandering third instar larvae began emerging. Homozygous females (purple) began emerging after seven days, while we observed a single homozygous male (blue) on Day 14. The expected number of larvae out of 190 for each sex and genotype is indicated with the grey background.
(D) The percent heterozygous and homozygous clamp1 and clamp2 adults that eclosed were counted from larvae collected in (B) and (C). While almost all heterozygous and homozygous clamp1 mutants eclosed, only the heterozygous clamp2 mutants eclosed.