Figure 5.
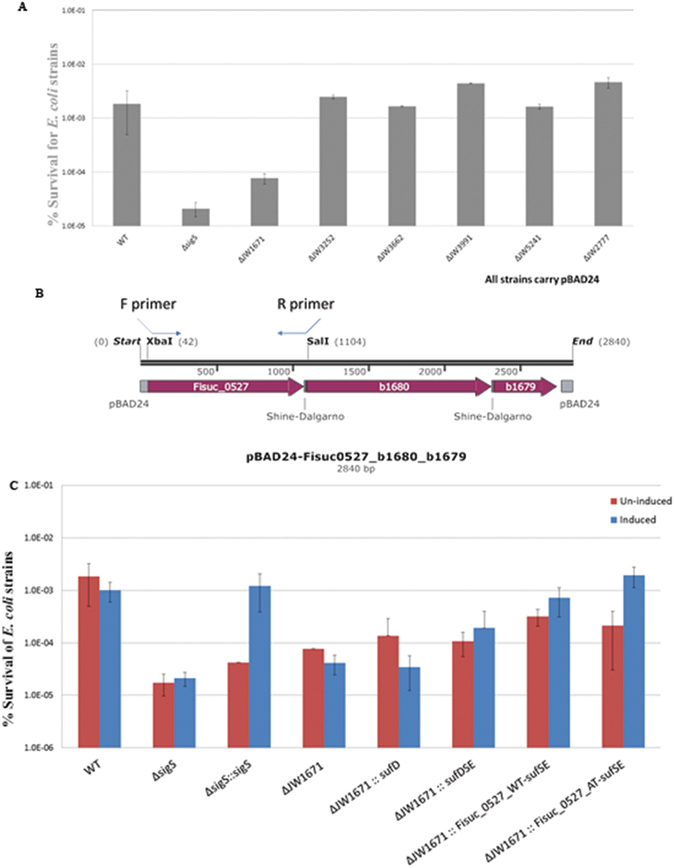
Acid survival assay of ΔJW1671 and complemented strains in E. coli. To investigate the roles of selected genes in the acid tolerant phenotype, E. coli strains deleted in individual genes homologous to acid tolerant F. succinogenes mutant genes were assayed using an acid survival CFU count method. A ΔsigS mutant was included as the acid sensitive control. (A) Among the six mutants, only the ΔJW1671 (sufD) mutant showed reduced ability to survival pH 2.5 treatment in minimal medium, compared to the wild type. (B) A linear map of the inset fragment of the fusion operon construct that was composed of F. succinogenes sufD fused with E. coli sufSE. A construct of sufDSE operon was first generated, and the sufD sequence was then replaced with the F. succinogenes counterpart. The reverse primer was designed to maintain the ribosome binding site of the immediate downstream gene. (C) Complementation of E. coli sufDSE but not sufD partially restored acid survival percentage, probably because the disruption of sufD caused a polar effect. Complementation with wild-type or acid tolerant F. succinogenes sufD restored the function of E. coli SufDSE, suggesting a possible role F. succinogenes sufD was involved in acid homeostasis.
